BME100 f2016:Group15 W8AM L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||||||||||||||||||||||||||||
|
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
have the same forward primer and reverse primer
do, the samples will become cross-contaminated
OpenPCR program After the test tubes are prepared, they are run through a thermal cycling machine. This machine heats and cools the test tubes several times, at various temperatures. Initially, the lid of the PCR machine is heated to 100°C, the boiling point of water. Heating this lid to 100°C will prevent the condensation of the water within the PCR test tubes, to the lid of the test tubes. Evaporation and condensation will remove the water from the PCR test tube, causing the reaction to fail. Once the lid is heated to 100°C, the test tubes are heated to 95°C for two minutes. This step prepares the DNA to be separated. After the DNA is heated to 95°C, the thermal cycling machine puts the PCR test tubes through a cycle of heating and cooling. The cycle involves 3 separate steps: Denaturing, Annealing, and Extending. During the denaturing process, the DNA is heated at 95°C for 30 seconds. It is this denaturing step that separates the DNA into two separate strands (Genome). The following step, annealing, cools the DNA to 57°C for 30 seconds. This step is where the primer attaches to beginning of the desired strand of DNA (Genome). The final step of the cycle, extending, heats the DNA to 72°C for another 30 seconds., and it is during this step in which the Taq DNA polymerase adds base pairs to the primer to create the new strand of DNA (Genome). This cycle of cooling and heating will be run 25 times, each time doubling the amount of DNA in the test tubes. Following the repetition of the cycle for a total of 25 times, the test tubes are cooled to 72°C for 2 minutes. This step allows for any partially completed copies to finish extending (Biolabs). The machine is then cooled to 4°C to maintain product integrity, and to allow for the test tubes to be removed. Works Cited Genome, Your. "What Is PCR (polymerase Chain Reaction)?" Yourgenome.org. The Public Engagement Team at the Wellcome Genome Campus, 25 Jan. 2016. Web. 18 Oct. 2016. Biolabs, New England. "PCR." Reagents For the Life Sciences Industry. New England BioLabs, n.d. Web. 18 Oct. 2016.
Research and DevelopmentPCR - The Underlying Technology
Polymerase Chain Reaction (PCR) targets a portion of DNA and makes millions of copies. PCR has applications in identifying bacteria, diagnosing diseases, and in other fields beyond medicine.
PCR includes four main components: template DNA, primers, Taq polymerase, and deoxyribonucleotides. The template DNA is the DNA containing the sequence meant to be amplified. Primers are short DNA sequences that bind to the Template DNA once it is split into two strands. They act as boundaries for the desired section of DNA during replication. These primers attract the Taq Polymerase enzymes responsible for replicating the DNA segment. Then, the Taq polymerase binds to primers and proceeds to replicate the desired section of DNA from in a 3’ to 5’ direction. The building blocks of this process are the deoxyribonucleotides cytosine, guanine, adenine, and thymine. These nucleotides converge with Taq Polymerase to continuously replicate the desired strand of DNA.
In PCR, the DNA, primers, polymerase, and deoxyribonucleotides are first mixed together in single test tube solution. The process for PCR then takes place over six steps of thermal cycling. First, it is heated up to 95 degrees Celsius for three minutes. This initial heating is used to isolate the actual template DNA from whatever matrix is came in be it cells within tissue or some bodily fluid. This is an important step to insure that no other reactions along with the PCR is taking place in during thermal cycling. At 95 degrees, the template DNA is split into two opposing strands as a result of increased molecular kinetic energy that weakens the hydrogen bonds between the original strands. This denaturing process takes place in about 30 seconds. The temperature is then lowered to 57 degrees for 30 seconds to allow DNA primers to bind to their corresponding regions along the template DNA strands. Finally, the temperature is raised to 72 degrees, which activates the DNA Taq Polymerase and enables it to bind to primers in preparation for replication and begin replicating the DNA strand in a 3’ to 5’ direction. This temperature is held for three minutes so that the polymerase can complete the assembling of nucleotides to replicated the desired DNA strand. In the base-pairing process, adenine (A) will anneal with base thymine (T), thymine with adenine, cytosine (C) with guanine (G), and guanine with cytosine. The last temperature change, a drop to 4 degrees Celsius, stops the activity completely to prepare for the repetition of another thermal cycle.
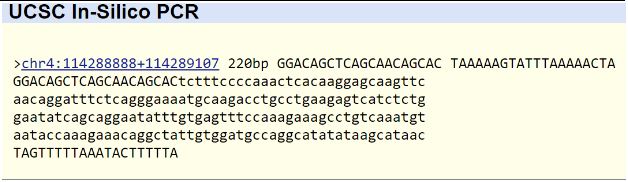
SNP Information & Primer DesignBackground: About the Disease SNP First off, SNP's are Single Nucleotide polymorphisms that are largely responsible for the genetic variation that exists among individuals. SNP's occur when a single nucliotide base pair varies between two genomes. This is caused by some sort of single point mutation like the deletion, insertion, or exchange of a single nucleotide. SNP's cause variations in genotypes and, if expressed, variations in phenotypes because they directly change the composition of proteins produced cells. DNA is transcribed into mRNA, which is translated in segments of three nucleotides called codons into specific amino acids that make up proteins. A single SNP can be responsible for altering the composition of a specific protein by changing the code for a particular amino acid. In this lab we are looking at the effects of a pathogenic SNP located on chromosome 4 section 113367751 that occurs in individuals that suffer from cardiac arrhythmia. This SNP is linked to cardiac arrhythmia because it causes the malfunction of ankyrin-2 (ANK2), a protien associated with ATPase binding, cytoskeletal adapter activity, and enzyme binding. The disease SNP causes the codon ATC to be translated, which embeds an amino acid in ANK2 that renders it non-functional and thus causes symptoms of cardiac arrhythmia. Primer Design and Testing Primers are meant to isolate the DNA segment containing the SNP we want to amplify. Therefore, the forward primer begins and contains the SNP and the reverse primer is found at the end of the DNA sequence of the SNP's respective protein, in this case ANK2. Taking that into consideration, the healthy forward primer was determined to be 20 nucleotides long starting from 113367751: 5'-GGACAGCTCAGCAACAGCAC-3’. The disease primer only differs in the last nucleotide, where the SNP is located, replacing cytosine with adenine: 5'-GGACAGCTCAGCAACAGCAA-3’. The reverse primer was designed based on the 20 nucleotides that follow the ANK2 gene: 5'-TAAAAAGTATTTAAAAACTA-3’. The disease reverse primer is the same as the healthy one because the reverse primer in this case is not associated with the SNP we are isolating. To make sure these primers would work in a PCR, meaning they would successfully bind to the DNA segment we want, a public record of the human genome was referred to. NCBI has a program called Primer-BLAST that finds matches between fabricated single-strand DNA sequences and ones that actually exist in the human genome. If there is a match, the program outputs the figure 220bp. So, we put in our healthy forward primer, and as seen in the image below, the program confirmed that the primer would work because it matched a specific sequence of DNA in the human genome. However, no matches were found when the disease forward primer was tested. This is normal considering the genome used in Primer-BLAST is that of a healthy individual who doesn't suffer from cardiac arrhythmia, and a disease primer would not match any sequence of DNA in a healthy individual.
| ||||||||||||||||||||||||||||||||