BME100 f2016:Group13 W8AM L6
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | |||||||||
|
OUR COMPANY
Arrow Dynamics---Where Accuracy is Everything LAB 6 WRITE-UPBayesian StatisticsOverview of the Original Diagnosis System
Calculations 1 and 2 serve to determine the reliability of the PCR and fluorimetry method in diagnosing for an SNP gene. Bayesian statistical analysis, in these two scenarios, is used to determine the probability that a patient will receive a positive conclusion, given that his or her DNA test for an SNP is positive, and the probability that a patient will receive a negative conclusion, given that his or her DNA test for an SNP is negative. The probabilities determined by both of these calculations was very close to 1.00 (100%), which implies that using PCR and fluorescence measurements is very reliable in determining whether a patient will receive a positive test conclusion given a positive diagnostic signal, and vice versa. Calculations 3 and 4 serve to determine the reliability of using PCR results to actually detect the development of an SNP-related disease. In these calculations, Bayesian analysis is used to determine the probability that a patient will develop the disease if he or she has a positive PCR conclusion, and the probability that a patient will not develop the disease if he or she has a negative PCR conclusion. The probabilities determined by these calculations were relatively low when compared with calculations 1 and 2, rounding up to about 0.7 (70%) for both calculations. This implies that the use of PCR and fluorimetry is only semi-reliable when tasked with determining whether a patient with a positive test conclusion will actually develop the disease, and vice versa.
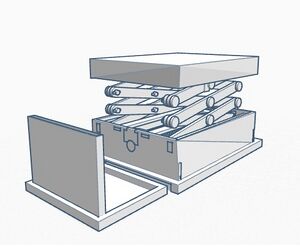
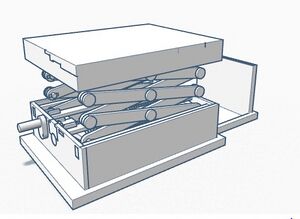
Sources of Error One possible human error that could greatly distort the Bayes values of the experiment would be the use of contaminated pipette tips to transfer samples from one microtube to another. This mistake would result in the mixing of each of the samples and would most likely skew the results of a group’s PCR reactions. A mechanical error that could negatively affect Bayes values is the stand used to orient the phone for photo-capturing. Our group found that it was difficult to properly keep the phone upright and stable at an angle suited for taking pictures of the DNA samples. If the pictures are analyzed in ImageJ but were taken from an awkward angle, the results of one’s measurements may end up skewed. A final mechanical error is the limitation of phone camera’s focal length when taking pictures of the DNA samples. Our group found it difficult to manually focus the camera on the drop we were placing on the slide, which is likely because of the short distance between the phone and the fluorimeter. Although we were able to obtain clear images from our experiment, blurry images caused by this mechanical error could affect a group’s ImageJ calculations and therefore negatively affect the Bayes values. Intro to Computer-Aided Design3D Modeling As a team we used Tinkercad to design our OpenPCR machine. Overall our experience was very positive in that the program itself was very intuitive and easy to use. At any point that we may have had a question on something there were tutorials along the way to help. The prefabricated designs provided by the professor also allowed for the whole design process to flow very smooth. One of the most important things that we as a team came to understand was simply how much goes into designing a machine such as this. Overall it was a very positive experience for us as a team to see and understand the work and thought that is needed to render our designs. Our Design
The design that we chose is ultimately the same structure as the original OpenPCR, however it will be smaller and incorporate pre-programmed settings to ensure accuracy of the PCR reaction.
Feature 1: ConsumablesWhile analyzing the consumables that we used for the lab during our OpenPCR experiment we concluded that while the products were easy to use and effective in conducting our experiment, we felt that the greatest weaknesses were the amount of waste produced and the possible room for error. During the original experiment, we had to pipette the buffer and SYBR Green solution into the individual OpenPCR test tubes. This process required repeated disposal of pipet tubes and the buffer and SYBR Green were both taken from larger tubes. This repeated in and out extraction left the experiment open to the possibility of cross-contamination.
Feature 2: Hardware - PCR Machine & FluorimeterPCR Machine While originally conducting the experiment ourselves we felt that the OpenPCR machine itself was easy to use, however, the programming for the device also allowed a certain room for error if it was not programmed correctly. In order to Our new OpenPCR design will be smaller and a new programming system will be applied to it. The smaller design will help the design and process be easier and quicker. The size will also make the product be more affordable to produce and for the consumer to purchase. The new programming system that will be applied to the new design will be able to eliminate any error in processing. This will ensure that the results are as accurate as possible.
| |||||||||