BME100 f2016:Group11 W8AM L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | |||||||||||||||||||||||||||||||||
|
OUR TEAM
LAB 4 WRITE-UPBold text==Protocol== Materials
Image Source:BME 100 Lab Workbook, Online
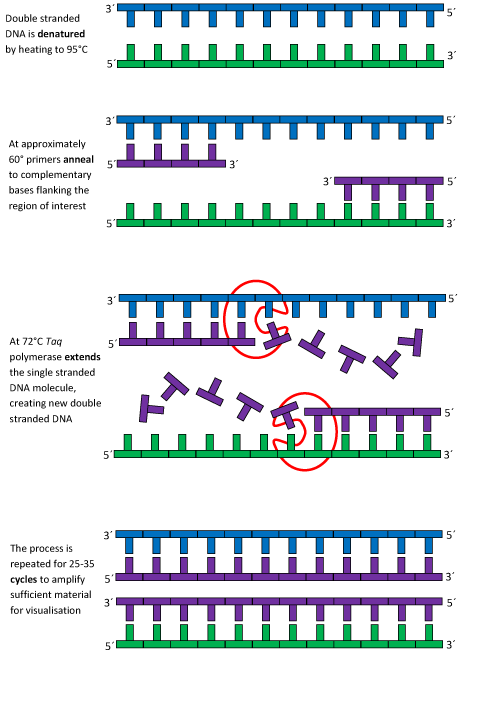
Research and DevelopmentPCR - The Underlying Technology Q1-What is the function of each component of a PCR reaction? The template DNA is the DNA being tested in the first place for a particular sequence within it. It will be denatured during the first step of the thermal cycling for detection of that particular sequence. Primers are short sequences of nucleotide, synthesized to bind, or anneal, to either end of the target sequence being replicated. Taq Polymerase is a protein which can function at high temperature that attaches to the complementary primers and extends a second strand of DNA, building nucleotides, off it as a base. Deoxyribonucleotides are constituent components of DNA which function as the "code" which is replicated with the taq polymerase to replicate the target sequence.
The initial step is to hold the mixture at 95 degrees Celsius for 3 minutes.During this step, the template DNA sample mixture is cleared of any contaminants by the high temperatures in order to ensure and prepare the PCR process for maximum efficiency. Next the template DNA denatures at 95°C for 30 seconds with the double-helix structure of the DNA separated into two strands as a result of the high temperatures. Annealing then occurs at 57°C for 30 seconds. During this point of the process, two short strands of DNA called primers anneal to their complementary matches on the separated DNA, on either end of the target DNA sequence that is to be replicated. The mixture is then extended at 72°C for 30 seconds where Taq polymerase binds to the primers and, using it as a starting point, forms nucleotide to further extend a newly formed second strand which comes out to be the target sequence. After the thermal cycling has been completed to satisfaction, the mixture is held at 72°C for 3 minutes. The purpose of this step is to extend any remaining primers missed by taq polymerase in previous thermal cycles to totally complete the process. Finally the the mixture is held at 4°C to prevent remaining DNA from replicating more and keep the result of the processes in a stable state for storing.
Q3: DNA is made up of four types of molecules called nucleotides, designated as A, T, C and G. Base-pairing, driven by hydrogen bonding, allows base pairs to stick together. Which base anneals to each base listed above? Adenine binds to Thymine with the reverse holding true and Cytosine binds to Guanine and vice-versa.
Q4: During which two steps of thermal cycling does base-pairing occur? Explain your answers. Base pairing occurs during the third and fourth steps, when annealing of primers and taq polymerase takes place receptively. In the former step, primers bind to separated strands of DNA and so base pairing must occur to bind the primers to that DNA. Taq polymerase in the fourth step extends nucleotides off those primers, extends a second strand of DNA and inevitably having to create a sequence of complementary nucleotides to bind onto the existing DNA strand.
Illustration of the PCR process. (2007, June 15). Retrieved October 17, 2016, from https://www2.le.ac.uk/departments/emfpu/genetics/explained/images/PCR-process.gif/view
SNP Information & Primer DesignBackground: About the Disease SNP SNP stands for "single nucleotide polymorphism." A nucleotide is a structural element of DNA and RNA. It consists of a base such as A, T, C, or G and a sugar with a phosphate. The polymorphism part means that there is genetic variation among the members of a community which leads to natural selection and survival of the fittest. The SNP variation is found in homo sapiens, which means humans. The chromosome that this change is found in is: 4:113367751. It is clinically significant, because it has a pathogenic allele. This change causes the cardiac arrhythmia syndrome, according to the citations. This SNP produces the protein, ANK2, which stands for ankyrin 2. Examples of the function of this protein are ATPase binding which is the selective interaction with an ATPase which itself is an enzyme that is a catalyst for the hydrolysis ATP. Another function is in its role in the binding of a particular molecule that allows the coordination of a cytoskeletal protein and one or more other molecules. ANK2 also plays an active part in enzyme binding where it non-covalently with any enzyme. An allele is one or more alternative forms of a gene that arises by mutation and are found on the same place on a chromosome. The disease associated allele for this nucleotide sequence contains the codon ATC instead of the 'correct' CTC codon. The numerical position of the SNP is 113367751.
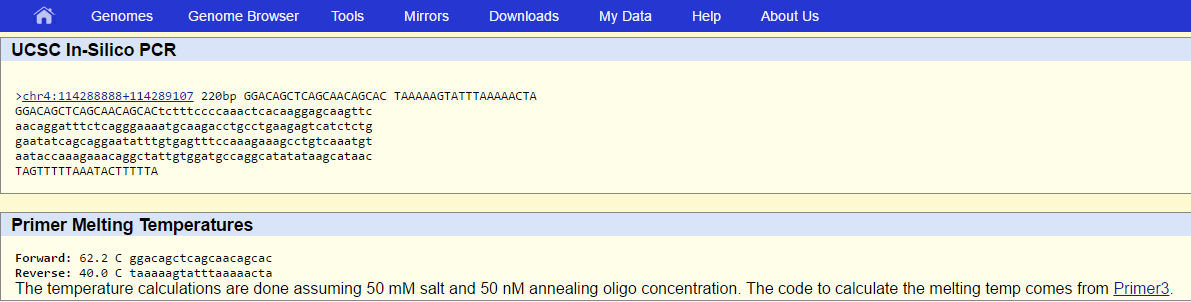
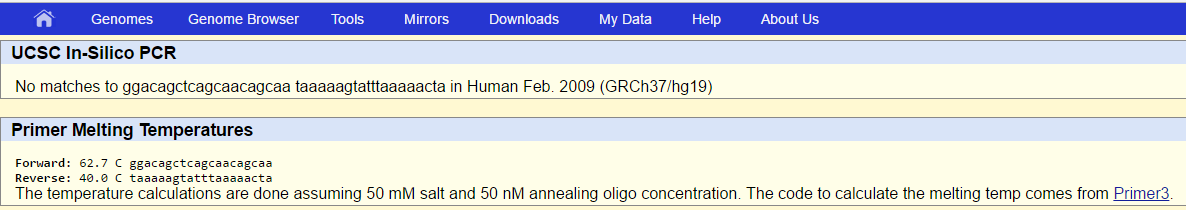
The primers that are specific to the SNP disease are below: The disease forward primer: 5'- GGACAGCTCAGCAACAGCAA - 3' Because in the SNP disease the part of the codon that is changed from normal A to C, in the primer that attaches to the following disease part also must be changed to C. Everything else is unchanged. The disease reverse primer: 5'- TAAAAAGTATTTAAAAACTA - 3'
Validation: Non-disease primer result:
Disease (SNP) primer result:
Kent, J. (2009). USCS In-Silico PCR. USCS Genome Bioinformatics. Retrieved from http://genome.ucsc.edu/cgi-bin/hgPcr?command=start.
The reason why the SNP primer is not in the database is because we are not looking up the normal DNA sequence (that showed up). We are looking up one of the polymorphisms. In other words, we are looking for a primer that is specific to one possible variation in the nucleotide out of 2 others. The 3 base variations are not normal; only 1 is the normal base that usually shows up in a majority of people before it is altered in a point mutation.
| |||||||||||||||||||||||||||||||||