BME100 f2014:Group9 L5
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | |||||||||||||||||||||||||||||||||||
|
OUR TEAM
LAB 5 WRITE-UPProcedureSmart Phone Camera Settings
Data AnalysisRepresentative Images of Negative and Positive Samples
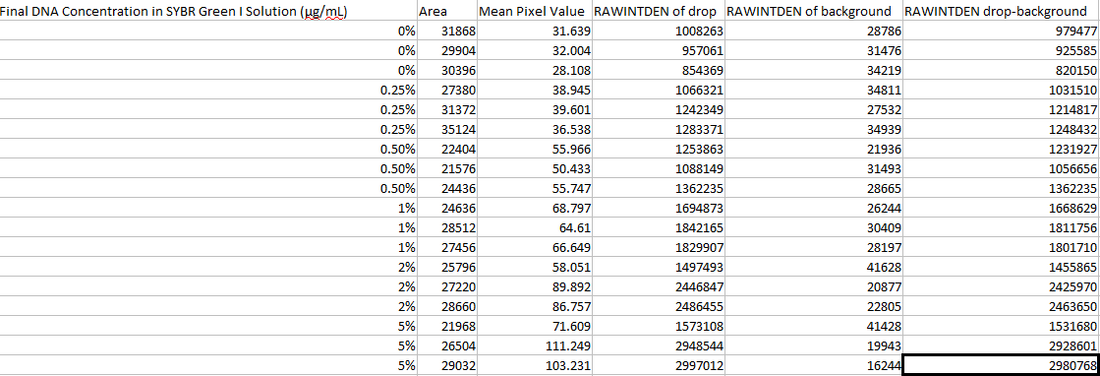
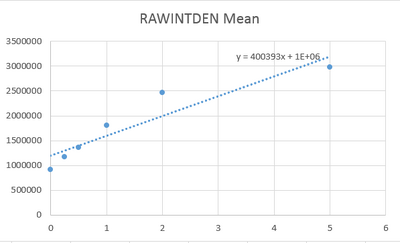
Image J Values for All Calibrator Samples Calibration curve
* Negative results may occur from error in the best-fit line Observed results
Conclusions
SNP Information & Primer DesignBackground: About the Disease SNP The disease SNP studied in this lab was categorized as rs16991654. It is a SNP found in the species homo sapiens, found in the chromosome 21:34370656. The clinical significance of this SNP is pathogenic, and it is associated with the KCNE2 gene, which has association with long QT syndrome, a rare heart condition. KCNE2 stands for potassium voltage-gated channel, Isk-related family, member 2. The function of this gene is to create a channel which functions in neurotransmitter release, heart rate, insulin secretion, neuronal excitability, epithelial electrolyte transport, smooth muscle contraction, and cell volume. It assembles with the product of another gene, which is a pore-creating protein, to alter its function.
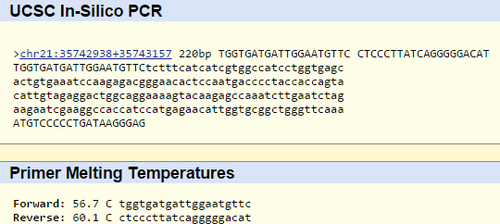
Primer Design and Testing In our primer test, we learned that the disease-associated allele contained the sequence CTC, and that the non-disease sequence was TTC. We used this information to find the forward primer for the non-disease sequence, which was TGGTGATGATTGGAATGTTC. 200 units from this was the reverse primer, which was CTCCCTTATCAGGGGGACAT. The disease forward primer was TGGTGATGATTGGAATGCTC, and the reverse primer was CTCCCTTATCAGGGGGACAT. These primers were taken to http://genome.ucsc.edu/cgi-bin/hgPcr?command=start, where we ran them through the program to determine the result. The non-disease primers finished with the result as follows: This validates our results in finding the correct primers, and when the test is run with the disease primers, no results are found.
| |||||||||||||||||||||||||||||||||||