BME100 f2014:Group8 L5
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | |||||||
OUR TEAM
LAB B/CPROCEDURECamera Settings:
Calibration: -The camera was placed 4 cm from the fluorimeter. The camera was aligned so that it was level with the fluorimeter. After this, a blackout box was placed over the camera and sample so that there was darkness when the photo was taken. Solutions Used For Calibration Part B To begin, each group was provided with three DNA samples, from two different patients. First, a positive control tube of DNA was produced by micropipetting 50 microliters of PCR reaction mix into an empty tube, then using a clean tip to micropipette the positive control DNA sample into the same tube. This yielded 100 microliters of solution. The same procedure was followed for the negative control DNA, and replicates 1, 2, and 3 for both patients 1 and 2. Then the tubes, each full of 100 microliters of PCR solution, were placed in the PCR heating block. The PCR cycles ran while the group cleaned up the equipment that was used, and the finished data would be acquired in Part C. Part C
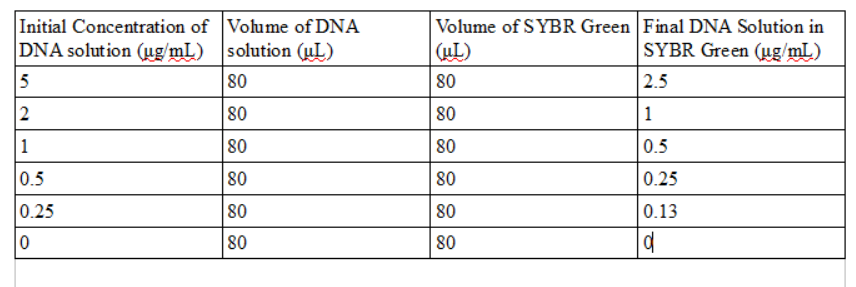
Next, 80 microliters of SYBR Green I solution was micropipetted onto the slide in the middle of the first two rows. Then 80 microliters of the calf thymus or water blank solutions were added to the drop. Once the drop was pinned on the slide, the slide was adjusted so that the blue LED light was focused by the drop onto the fiber optic fitting on the other side of the drop. The timer on the group's camera could not take a picture without creating a flash, so one team member had to take pictures with their hand inside the light box, preventing as much light as possible from entering and interfering with the image. Three images of each drop were taken. The pipettor was used to remove all 160 microliters of solution from the slide, and the procedure was repeated for each concentration of calf thymus DNA given by the instructions. In continuation of the PCR reactions which were set up in part B, the PCR samples were obtained and measured using the fluorimeter. The micropipettor was set to 120 microliters and all 100 microliters of PCR sample were transferred into the tubes which contained buffer solution. This procedure was followed for the rest of the PCR sample tubes as well. Next, 80 microliters of the PCR solution which had been diluted by the buffer solution was pipetted onto a slide, along with 80 microliters of SYBR GREEN I, and again, three images of each drop were captured. All tubes were disposed of in hazardous waste bags, and equipment was returned after fluorimeter analysis was finished.
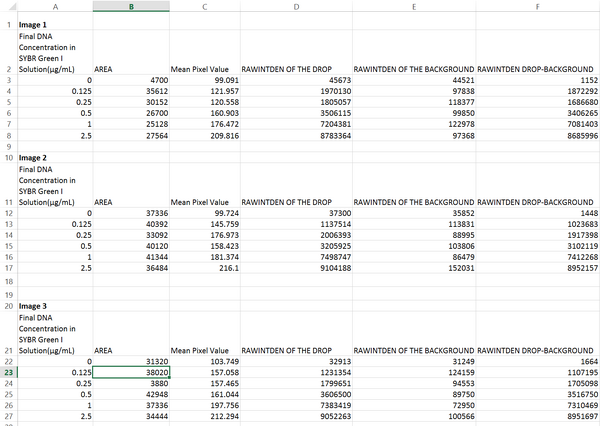
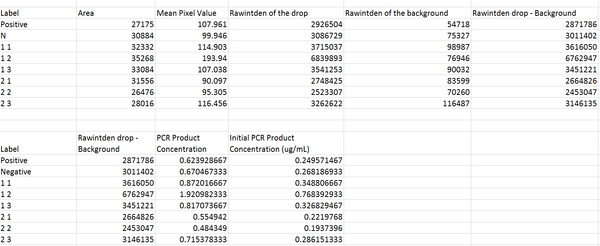
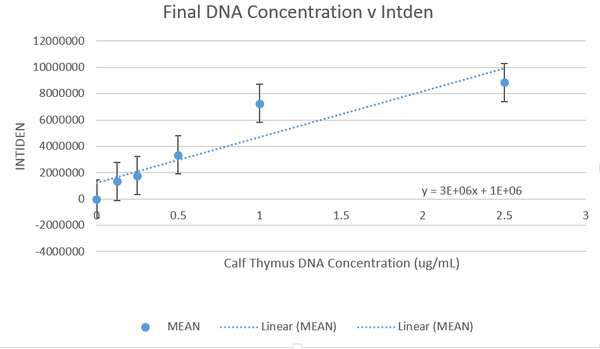
DATA ANALYSISImage J Values Data Summary:
LAB DSNP INFORMATION AND PRIMER DESIGNPart 1- Vocabulary-
rs16991654-
-The rs16991654 variation is found in Homo Sapiens.
-It’s located on the chromosome 21
-The clinical significance is pathogenic.
-It’s associated with the KCNE2 gene.
-The rs16991654 variation is linked to Long QT syndrome. Part 2-
-Potassium voltage-gated channel, Isk-related family, member 2
-This gene regulates the movement of potassium ions in and out of a system. It affects neuron function, heart function, pancreatic eyelet cells, and tissue covering.
-Set of nucleotides which affect the development of proteins and phenotypic variances in organisms.
-CTC
-34370656 Part 3-
- 5'-CATGGTGATGATTGGAATGT
-34370856
- 5'-CCCTTATCGAGGGGGACATT
- 5'-CATGGTGATGATTGGAATGC
- 5' -CCCTTATCGAGGGGGACATT |
|||||||