BME100 f2014:Group3 L5
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | |||||||||||||||||||||||||||||||||||
|
OUR TEAM
LAB 5 WRITE-UPProcedureSmart Phone Camera Settings
Solutions Used for Calibration
This picture shows our smartphone resting in the phone cradle, 4.5 cm away from the slide where the drop is placed. This picture shows our setup in the black box that will be used to block out light. Once the camera's timer is set off, the front flap of the box will be closed so no light interferes with the picture.
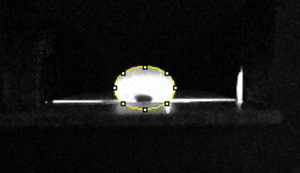
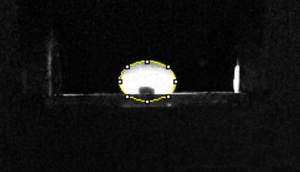
Data AnalysisRepresentative Images of Negative and Positive Samples
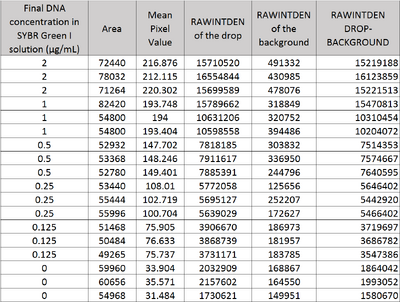
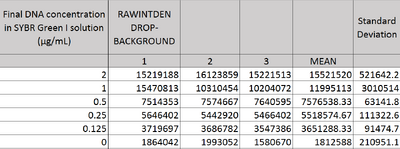
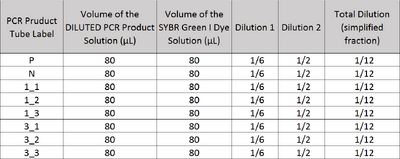
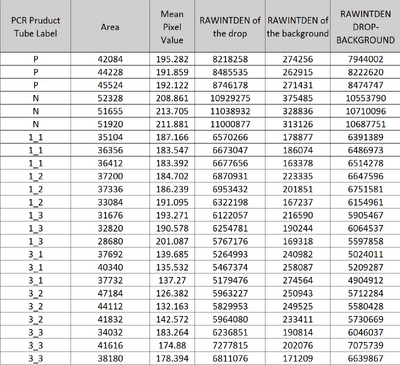
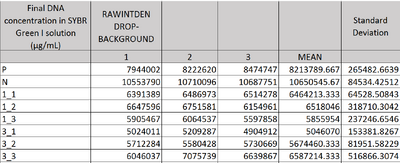
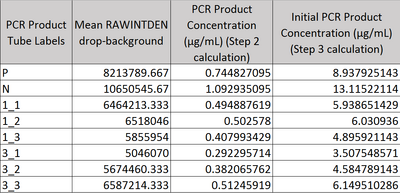
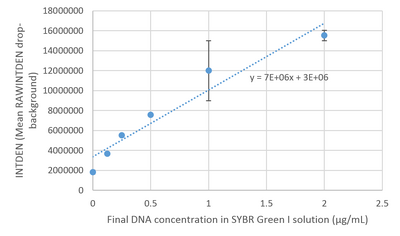
Image J Values for All Calibrator Samples Additional Tables used for Calculations
Observed results
Conclusions
We were unable to draw conclusions about either of our patients because there was an error in our negative control. It appears that somewhere during the experiment's course the negative sample was contaminated with a positive strain of DNA because our negative control drop glowed green, which it should not have done. Because of this error, we are unable to draw any conclusions from our data.
SNP Information & Primer DesignBackground: About the Disease SNP A nucleotide/s is structural components of DNA and RNA (series of building blocks if you will). DNA and RNA assembled of individual nucleotides. Nucleotides are consisting of three organic molecules; a 5 carbon sugar (Deoxyribose or ribose), a phosphate group and one of four set of nitrogenous base (A, T, C and G).Poly means two or more; morphs means, slow variation within a same species. Therefore, polymorphism is an existence of two or more distinct/obvious different phenotype among the populations of a same species (genus if you will). It is found in the Homo Sapiens species. The chromosome variation is located on; 21:34370656. Of this SNP the clinical significance is pathogenic. The gene associated with SNP is KCNE2. The disease associated with this SNP is congenital long QT syndromes (LQTSs).
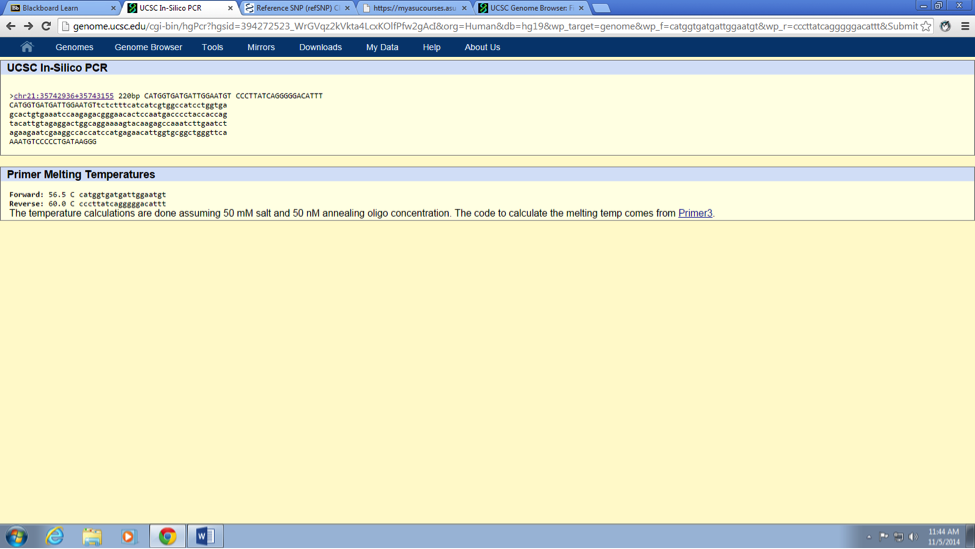
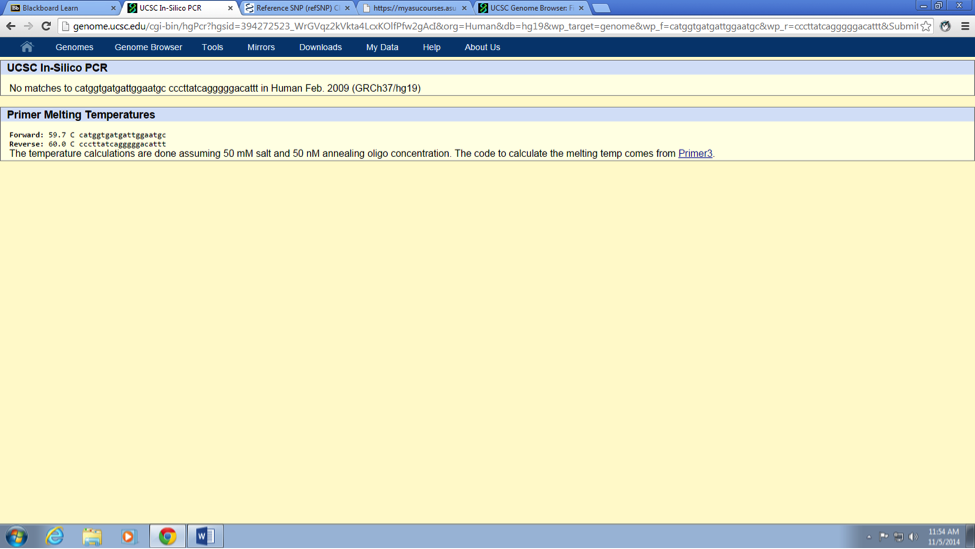
What does KCNE2 stand for? Potassium voltage-gated channel, Isk-related family, member 2 Briefly describe the molecular function of this gene: The KCNH2 gene in human sapiens DNA, codes for a protein that rise to a potassium ion channel. This potassium ion channel is responsible to regulate electrical current activity of the heart beats. If for some reason (such as mutation in a single nucleotide), the functionality of this potassium ion channel inhibited or changed (i.e. lost or gain functionality of ion channel) it may cause a mortal disorder know as long QT syndrome. Patients associated with long QT syndrome, have a potential risk of fatal cardiac arrhythmias or sudden death. What is an allele? An alternative form or different version of a gene, called an allele. Furthermore, these diverse alleles may rise to different phenotypic traits. The disease-associated allele contains what sequence? CTC The numerical position of the SNP is: 34370656 Non-disease forward primer (20 nt): CATGGTGATGATTGGAATGT The numerical position exactly200 bases to the right of the disease SNP is: 34370856 Non-disease reverse primer (20 nt): CCCTTATCAGGGGGACATTT Disease forward primer (20 nt): CATGGTGATGATTGGAATGC Disease reverse primer (20 nt): CCCTTATCAGGGGGACATTT | |||||||||||||||||||||||||||||||||||