BME100 f2014:Group1 L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||||||||||||||||||||||||||||||
|
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
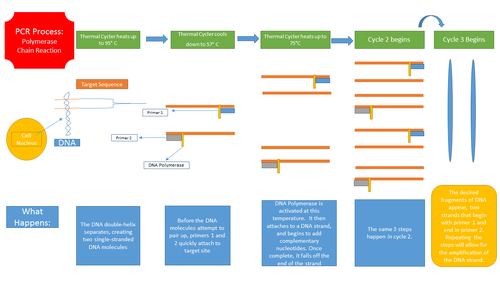
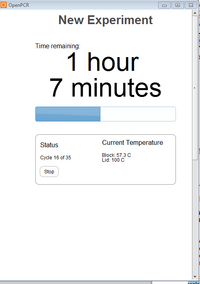
Polymerase Chain Reaction Process Resulting in the Amplification of a single DNA strand OpenPCR program The thermal cycling program is used to amplify segments of DNA via the PCR. It heats and cools the DNA in cycles. It has two parameters including Primer Annealing as well as Primer Extension. Primer Annealing depends on the length, base composition, as well as concentration of the amplification primers. Extension depends upon the length and concentration of the target sequence and temperature. The optimum number of cycles depends mainly on the starting concentration of the target DNA. The optimum temperature for denaturation is about 90-95 degrees Celsius.
Research and DevelopmentPCR - The Underlying Technology Template DNA within a PCR reaction is copied or amplified throughout the process in order to create billions of identical, specific DNA sequences that can be used to diagnose diseases, identify people based on DNA (i.e. criminals or relatives), or to identify different types of viruses or bacteria. In the PCR reaction as the DNA is duplicating, a primer, short piece of laboratory made DNA, binds to the template DNA in order to start the copying process. Taq Polymerase, a complex of proteins that occurs naturally and copies the DNA before the DNA divides in two, attaches to the end of the primers and starts to add nucleotides. The deoxyribonucleotides are four "building blocks" that make up DNA molecules. They come in four different types and A, C, G, and T. There purpose is to store and transmit the genetic information to form the duplicate DNA in the PCR reaction. Initial Step for 3 minutes: 95°C Ensures all template DNA is denatured into two single stranded DNA molecules. 35 cycles of: 95°C 30 seconds of Denaturation: The templete DNA is split into two single stranded DNAmolecules. 57°C 30 seconds of Annealing: The forward and reverse primers bind to their appropriate nucleotide sequences on both single stranded DNA molecules. 72°C 30 seconds of Extension: Taq polymerase binds to the primer and begins to add corresponding nucleotides(dNTPs) to the single stranded DNA strand creating two double stranded DNA for each one DNA strand that we began with. Final Step 72°C for 3 minutes: Continues extension to ensure that all single stranded DNA is fully extended. Final Hold 4°C: To ensure that bonding occurs within the strands of DNA. Pair molecules: A(Adenine) binds to T(Thymine) G(Guanine) binds to C(Cytosine)
| ||||||||||||||||||||||||||||||||||