20.109(S15):DNA sequencing (Day5)
Introduction
Last time you reacted your 16S PCR product with specially prepared backbone DNA, and then transformed the reaction product into an engineered cell strain. Eight independent colonies were selected from each of your plates and grown overnight in liquid culture. You will now isolate and sequence DNA from each colony, then pool your results with those from other students studying that particular bird sample to construct a phylogenetic tree representing the bacterial composition in that sample.
We have already discussed the features of the transformation strain needed to synthesize and select for the whole plasmid, namely the ccdB gene. Now let's talk about those features relevant to extracting DNA. As you can gather from the linked manual (PDF), the One Shot TOP10 cells have endA and recA mutations, both of which make cloning go more smoothly. First, endA1 limits the non-specific destruction of plasmid (and chromosomal) DNA normally carried out by the EndA enzyme, thus maximizing DNA recovery. (The cells also have additional deficiencies in restriction-based nucleases.) Second, recA1 makes the cells incapable of homologous recombination, which could otherwise cause undesirable intermingling between the plasmid and chromosomal DNA.
The procedure for DNA isolation at this scale is commonly termed "mini-prep," which distinguishes it from a “maxi-prep” that involves a larger volume of cells and additional steps of purification. The overall goal of each prep is the same--to separate the plasmid DNA from the chromosomal DNA and cellular debris, allowing the plasmid DNA to be studied further. In the traditional mini-prep protocol, the media is removed from the cells by centrifugation. The cells are resuspended in a solution that contains Tris to buffer the cells and EDTA to bind divalent cations in the lipid bilayer, thereby weakening the cell envelope. A solution of sodium hydroxide and sodium dodecyl sulfate (SDS) is then added. The base denatures the cell’s DNA, both chromosomal and plasmid, while the detergent dissolves the cellular proteins and lipids. The pH of the solution is returned to neutral by adding a mixture of acetic acid and potassium acetate. At neutral pH the SDS precipitates from solution, carrying with it the dissolved proteins and lipids. In addition, the DNA strands renature at neutral pH. The chromosomal DNA, which is much longer than the plasmid DNA, renatures as a tangle that gets trapped in the SDS precipitate. The plasmid DNA renatures normally and stays in solution, effectively separating plasmid DNA from the chromosomal DNA and the proteins and lipids of the cell.
Normally in 20.109 we do an in-house mini-prep procedure according to the steps above followed by ethanol precipitation. However, because you are isolating DNA from so many colonies, today we will use a commercially available kit so that the work can go more quickly. The principle is the same as that of our "quick and dirty" (and cheaper!) prep, but is combined with the silica gel column purification you are familiar with from using other Qiagen kits.
After isolation, you will quantify your DNA by spectrophotometry. Nucleic acids (both RNA and DNA) have an absorbance peak at 260 nm. Beer's law may be used to quantify the amount of DNA from this peak: Abs = ε l c, where Abs is the measured absorbance, l is the path length (1 cm for most specs), c is concentration, and ε is the extinction coefficient. For DNA, ε is 0.02 (μg/mL cm)-1, so 1 absorbance unit corresponds to 50 μg/mL of DNA. The absorbance at 280 nm gives some indication of DNA purity, as proteins have their absorbance peaks at that value (primarily due to the aromatic peptides tryptophan and tyrosine). An Abs260:Abs280 ratio of ~1.8:1 is desired.
Miniprepped DNA will be sent for sequencing off-site. Each clone will be sequenced from two directions, in order to cover the entire 1400 bp region of interest. Together, you'll do about 200 sequencing reactions per lab section!
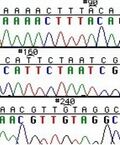
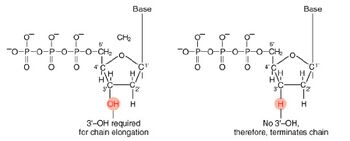
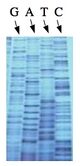
The invention of automated sequencing machines has made sequence determination a relatively fast and inexpensive endeavor. The method for sequencing DNA is not new but automation of the process is recent, developed in conjunction with the massive genome sequencing efforts of the 1990s. At the heart of sequencing reactions is chemistry worked out by Fred Sanger in the 1970s which uses dideoxynucleotides (see schematic above left). These chain-terminating bases can be added to a growing chain of DNA but cannot be further extended. Performing four reactions, each with a different chain-terminating base, generates fragments of different lengths ending at G, A, T, or C. The fragments, once separated by size, reflect the DNA’s sequence. In the “old days” (all of 20 years ago!) radioactive material was incorporated into the elongating DNA fragments so they could be visualized on X-ray film (image above center). More recently, fluorescent dyes have been used instead, with one color linked to each dideoxy-base. The four colored fragments can be passed through capillaries to a computer that can display the color intensities detected (image above right).
Today you will also screen bird samples for AIV using the primers you designed on Day 2. Due to the potential risk of working with bird samples that contain AIV, Wendy Puryear from the Runstadler Lab isolated RNA from bird cloacal samples then the teaching faculty generated the cDNA that you will use for your assay. You will be using cDNA because it is more stable than RNA. As mentioned previously, your screen will employ RRT-PCR. This technique builds upon standard PCR by enabling researchers to quantitatively determine the amount of template present in a complex sample. For example, it is possible to calculate the number of transcripts for a specific gene within a sample.
For your screening assay, you will use the threshold cycle (CT) to assess the sensitivity of your primers. The threshold in RRT-PCR corresponds to a fluorescence measurement that is slightly above background and the cycle at which this occurs for a given sample is the CT. On Day 8 we will discuss the specifics of RRT-PCR and CT in preparation for your primer design memo assignment.
In another week you will finally get to see the results of all your hard work!
Protocols
Part 1: Extract DNA from selected clones (mini prep)
Last year most folks chose to prepare all 16 candidates at once, and this timing worked well in the main. However, there were definitely bottlenecks at both the spec. and the sequencing plates, which might be reduced somewhat if a few groups try the suggestion below.
Each team today has sixteen minipreps to do. You may find it easier to complete these in two shifts rather than to attempt to quickly pipet across all sixteen samples. One approach you could take is the following: start with the first eight candidates (A group), then spin down the next eight (B group) during the 5 min lysis step for the A group; finally, start adding Soln. I to the B group when you reach the 10 min spin step for the A group. As one partner wraps up the B group, the other could begin Part 4.
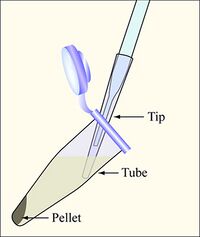
- Pick up your sixteen candidates cultures, which are growing in the test tubes labeled with your team color. Label eight eppendorf tubes to reflect each partner's candidates (Rd1-8 and Rd9-16).
- Vortex the bacteria and pour ~1.5 mL of each candidate into an eppendorf tube.
- Balance the tubes in the microfuge, spin them at maximum speed for two minutes, and remove the supernatants with the vacuum aspirator, as shown in the diagram. Add a fresh 200uL non-filtered pipette tip on the glass aspirator, and change 200uL tips between each sample to avoid cross-contamination.
- Pour another 1.5 mL of the same culture onto the pellet, and repeat the spin step.
- Resuspend the cell pellet in 250 μL Solution I.
- Soln. I contains RNase so that we collect only our nucleic acid of interest, DNA.
- Mix thoroughly by pipetting or vortexing.
- Add 250 μL of buffer Solution II and mix by gently inverting or rotating the tube about 4-6 times. You may incubate here for up to 5 minutes, but not more.
- Soln. II contains sodium hydroxide for lysing.
- Add 350 μL buffer Solution III, and mix immediately by inversion.
- Soln. III contains acetic acid, which will cause the chromosomal DNA to messily precipitate; the faster you invert, the more homogeneous the precipitation will be.
- Soln. III also contains a chaotropic salt in preparation for the silica column purification.
- Centrifuge for 10 minutes at maximum speed. Note that you will be saving the supernatant after this step.
- Meanwhile, prepare 8 labeled HiBind DNA columns per person, one for each candidate clone, and 8 trimmed eppendorf tubes per person for the final elution step. Label and save your eppendorf lids for the final elution step.
- Place each HiBind column over a 2 mL collection tube, add 100 μL of 3M NaOH to equilibrate, and centrifuge for 30 seconds. The NaOH can be dumped right in the sink.
- Transfer the entire supernatant to the pre-equilibrated column and centrifuge for 1 min. Discard the eluant into a temporary waste tube.
- Next wash with 0.5 mL HBC, with a 1 min spin step as usual. Discard the eluant as before.
- Now add 0.7mL DNA Wash Buffer, with a 1 min spin step as usual. Discard the eluant as before.
- This buffer contains a high concentration of ethanol to precipitate the DNA.
- After removing the DNA Wash Buffer, spin the mostly dry column for 1 more min.
- It is important to remove all traces of ethanol, as they may interfere with subsequent work with the DNA.
- Transfer the column to your trimmed eppendorf tube. Add 50 μL of sterile water, pH 8.5, to the top center of the column, wait 1 min, and then spin 1 min to collect your DNA.
Part 2: Count colonies
When you have a moment today, count the colonies on your plate from last time and post the values on today's Talk page. Don't forget to count the colonies we have already picked!
Part 3: Measure DNA concentration
You will spot-check 3 of 8 minipreps per person before preparing sequencing reactions. If all are reasonably similar, you can then choose an average or threshold value of DNA volume that should work for all sequencing reactions. For example, you might use 2 μL in each reaction for simplicity, rather than sometimes 1 μL, other times 2.5 μL, etc. The reactions should be robust within a few-fold DNA concentration.
- For each DNA sample that you measure, you will combine 6 μL of miniprepped DNA with 474 μL of water. This dilution ought to give you a measurement that's in a reliable range. You can prepare your DNA dilutions directly in cuvettes and mix them well.
- The best approach might be to add the DNA first, then add the water and pipet up and down to mix. Avoid making bubbles or losing volume.
- Notice that today we are using cuvettes made of a special plastic that is transparent to UV light.
- To get the most accurate measurement, you should also prepare a blanking solution that contains everything except the DNA, i.e., 474 μL of water and 6 μL of elution buffer (water!).
- At the spec, enter Nucleic Acid Analysis mode. Option 4 should be highlighted, and you can hit START to proceed.
- Begin with the cuvette containing blanking solution, and hit Blank on the spectrophotometer. As you proceed, be sure to press each cuvette all the way down.
- Proceed to take an absorbance scan of each DNA sample. Record the 260 nm and 280 nm absorbance values in your notebook. The spec will calculate the purity ratio for you, but not the DNA concentration.
- What assumption are we making about the cuvettes when we put the blanking solution in a separate cuvette from the samples?
- Calculate the eight DNA concentrations, along with the volume of DNA required to use 500 ng in a sequencing reaction. You may find the table below helpful.
- Recall that an absorbance value of 1 indicates a concentration of 50 μg/mL of the measured DNA (i.e., the diluted solution and not the original stock).
- Which of the two wavelengths indicates DNA quantity? Re-read the introduction if you're not sure.
- If your samples require adding less than 1 μL of DNA, prepare intermediate dilutions first. For example, instead of adding 0.9 μL of stock, you could add 9 μL of a 1:10 dilution of that particular miniprep. No sequencing reaction should contain more than 10 μL of DNA.
- Recall that an absorbance value of 1 indicates a concentration of 50 μg/mL of the measured DNA (i.e., the diluted solution and not the original stock).
| # | A260 value | A280 value | Ratio | Measured Conc. | Stock Conc. | Volume for 500 ng DNA | Volume H2O (see below) |
|---|---|---|---|---|---|---|---|
| 1 | |||||||
| 2 | |||||||
| 3 | |||||||
| AVE |
Part 4: Prepare sequencing reactions
As we will discuss in lab today, sequencing reactions require a primer for initiation. Legible readout of the gene typically begins about 40-50 bp downstream of the primer site, and continues for ~1000 bp at most. Thus, multiple primers must be used to fully view amplicons > 1 Kbp in size, such as your ~ 1400 bp 16S sequence. We will use forward and reverse primers that anneal to the vector upstream and downstream of the 16S insert in order to capture the entire sequence for analysis.
The recommended composition of sequencing reactions for this type and length of DNA is ~500 ng (though 200 ng should be plenty in a pinch) and 25 pmoles of sequencing primer. Genewiz will add our primers for us (5 μL / reaction)!! Each of your sequencing reactions should thus consist of 13 μL of aqueous DNA (3 μL extra to account for pipetting error). For example, if you are adding only 2 μL of DNA for a particular candidate, you must add another 11 μL of water to complete the sequencing mixture. Genewiz will take 10 μL of your DNA + water and transfer it to a plate with 5 μL of their primer. The final sequencing reaction volume is 15 μL.
Each team will fill 4 columns on a 96-well plate, according to the table below. Odd-numbered columns will contain forward primer (added later by Genewiz) and even-numbered columns should contain reverse primer (added later by Genewiz). You can add pre-mixed DNA/water preparations individually to wells (be sure to make enough of each clone for both forward and reverse primer testing!). Per team, the first two columns can contain C1-C8, and the next two C9-C16. After you have finished distributing each sample, cover that column with a strip of caps.
| Team color (T/R) | Plate # | Column #s | Team color (W/F) | Plate # | Column #s |
|---|---|---|---|---|---|
| Orange | 1 | 1-4 | Red | 1 | 1-4 |
| Yellow | 1 | 5-8 | Orange | 1 | 5-8 |
| Green | 2 | 1-4 | Yellow | 2 | 1-4 |
| Blue | 2 | 5-8 | Green | 2 | 5-8 |
| Purple | 2 | 9-12 | Blue | 3 | 1-4 |
| Pink | 3 | 5-8 | |||
| Purple | 4 | 1-4 | |||
| Silver | 4 | 5-10 |
Homework
Due on M1D6
- Some of you have journal club presentations next time. No other homework is due on Day 6.
Due on M1D7
- One week from today you will submit an outline of the background and motivation section of your data summary. Feel free to read the assignment details and get an early start!
Reagent list
- E.Z.N.A Plasmid DNA Mini Kit I (Omega Bio-Tek)
- Sequencing primers (5 μM stocks)
- M13 (F) sequence: CAGGAAACAGCTATGAC
- T7 (R) sequence: GTA ATA CGA CTC ACT ATA GG
- 2X Power SYBR Green from Applied Biosystems
- RRT-PCR primers (20 μM stocks)
- cDNA (1 ng/μL)
Navigation Links
Next Day: Journal club I Previous Day: DNA cloning