20.109(S13):PCR and paper discussion (Day3)
Introduction
Last time you prepared a DNA pool from a bird stool sample, and today you will amplify 16S rRNA sequences from that pool using PCR. Recall that PCR consists of repeated melting, annealing, and extension steps. During the annealing step of PCR, primers should largely bind to the target sequence, but some may bind to off-target sequences as well. Specificity of binding is controlled by both primer design and reaction conditions.
During the reaction itself, we can improve the reliability and accuracy of PCR by two key methods. The first is the use of a highly specific polymerase, one with either engineered or inherent hot start properties. Hot start means that the polymerase is inactive at low temperatures. Polymerases officially called "hot start" are negligibly active at room temperature, allowing reaction assembly without chilling, and also exhibit relatively lower activity at typical annealing temperatures (50-55 °C) than other polymerases do. These properties tend to reduce binding of primers to non-target DNA. The polymerase you will use has much activity at typical annealing temperatures than does the laboratory workhorse Taq, but is not officially a hot start polymerase. One important point to note here is that the target DNA may be present at a low concentration compared to the DNA in the sample as a whole, especially in complex mixtures, thus increasing opportunities for non-specific binding.
The second performance enhancer we will use is bovine serum albumin (BSA), which is an especially important additive for amplifying DNA originating from a stool sample. Recall that stool samples are replete with inhibitors, including those that bind directly to DNA polymerases. As you saw if you clicked on the Kreader paper linked on Day 2, BSA itself binds many inhibitors of PCR, thus acting as a competitor. We would much rather that inhibitors bind the BSA than bind the polymerase and interfere with its function! BSA is hydrophobic and somewhat positively charged, making it a great non-specific binder of proteins that we will use time and again in 20.109.
Next time, you will visualize your entire reaction mixture in a gel, and if need be excise and purify the band of the correct size (~1400 bp) to isolate it from any non-specific products that occur in spite of the above precautions. Gel electrophoresis is a technique used to separate large molecules by size using an applied electrical field and appropriate sieving matrix. DNA fragments are typically separated in gels composed of agarose, a seaweed-derived polymer (see figure, below left). To prepare these gels, molten agarose is poured into a horizontal casting tray containing a comb. Once the agarose has solidified, the comb is removed, leaving wells into which the DNA sample can be loaded. The loaded DNA samples are then pulled through the matrix when a current is applied across it. Specifically, DNA molecules are negatively charged due to their phosphate backbones, and thus travel toward the positive charge at the far end of the gel (see figure, below right).
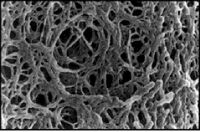
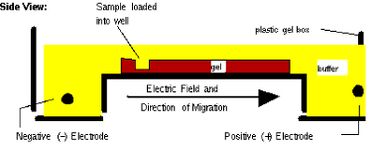
Although all DNA molecules travel in the same direction during gel electrophoresis, they do so at different rates: larger molecules get entwined in the matrix and retarded, while smaller molecules wind through the matrix more quickly and thus travel further from the well. Ultimately, fragments of similar length accumulate into “bands” in the gel. Bands of DNA are usually visualized by adding the fluorescent dye ethidium bromide (or newer alternatives such as SYBR Safe) to agarose gels. This dye intercalates between the bases of DNA, allowing DNA fragments to be located in the gel under UV light and photographed. The intensity of the band reflects the concentration of molecules that size, although there are upper and lower limits to the sensitivity of dyes. Because of its interaction with DNA, ethidium bromide is a powerful mutagen and will interact with the DNA in your body just as it does with any DNA on a gel. You should always handle all gels and gel equipment with nitrile gloves. Agarose gels with ethidium bromide must be disposed of as hazardous waste.
One parameter that affects the way DNA travels through a gel is the pore size, which is in turn affected by both the weight percent of the gel and the type of agarose used. Because we are separating large DNA fragments (> 1 Kbp) in the bacteria experiment, a low-to-medium percentage (namely 1.5 %) gel is appropriate. In the microsporidia experiment, small fragments (~ 0.1 Kbp) are expected and thus a high percentage (namely 3%) gel will be used. Moreover, we will use high-resolution (HR) agarose; its low viscosity means that high weight percent solutions are tractable to work with, and that the solidified gel remains pliable rather than brittle. HR agarose can be prepared by chemically modifying and/or partially depolymerizing natural agarose (as described here).
The PCR will continue during the whole lab period. In the meantime, we will discuss a journal article, both to learn more about investigations of the microbiome and to become comfortable reading and discussing the primary scientific literature. You will also hear from our oral presentation instructor on how to give a good talk, and get immediate feedback on your informal presentation of one slide together with your partner. In 2-3 weeks, you will each present an article on your own.
Protocols
Part 1: Prepare PCR to detect bacterial 16S
- Begin by carefully labeling each PCR tube that you will use with the date, sample name, and a unique symbol and/or color for quick identification. Filling in the cap tab with your team color will usually suffice.
- Pre-chill the tubes on a cold block.
- You and your partner can now prepare and share a so-called "master mix," which contains every PCR ingredient except the template and the polymerase. Prepare enough for the number of reactions you need to run, plus an additional 10%. In addition to the two DNA-containing reactions, you should prepare a no-template control that contains pure water without any plasmid. Feel free to use the table below for your calculations.
- When the master mix is not in use, keep it on ice.
- What do you expect to see in the no-template control case?
- Combine 45 μL of master mix, 5 μL of template, and 1 μL of PfuUltra polymerase in a PCR tube. When everyone's reactions are ready, they will undergo the cycling conditions listed below.
- Add the master mix first, because the template alone may freeze. Then add template and polymerase, and finally (gently!) mix the reaction with a larger pipet.
| Reagent | Amount for 1 reaction (μL) | Amount for 3 reactions + 10% |
|---|---|---|
| PfuUltra buffer (10X stock) | 5 | |
| Primer mix | 1 | |
| dNTPs | 1 | |
| 5% BSA (100X stock) | 0.5 | |
| Water | 37.5 | |
| DNA template | 5 | N/A |
| Segment | Cycles | Temperature (° C) | Time |
|---|---|---|---|
| 1 | 1 | 95 | 5 min |
| 2-4 | 35 | 95 | 1 min |
| 51 | 1 min | ||
| 72 | 2 min | ||
| 5 | 1 | 72 | 10 min |
| 6 | 1 | 4 | indefinite |
Parts 2 + 3: Journal article discussion and WAC session
Scientific papers are dense and often time-consuming to read and understand, but with practice, you will find strategies that improve your comprehension efficiency. Here's one tip to get you started: when reading newly reported results, be sure to refer to the associated figures frequently, because visual information is often easier to take in than purely verbal descriptions.
Technical Background
Several terms and approaches in the Koenig et al. paper may be unfamiliar to you. Here we will provide some background on selected topics. You should feel free to search for information about additional topics -- even Wikipedia can be a good start! At the same time, don't feel the need to understand every detail presented in the paper.
Named for the company that developed the technique, 454-pyrosequencing is one example of a next generation sequencing method. Such methods were designed to enable efficient and accurate identification of many, many sequences at once. You can learn about this particular approach at the company website.
Metagenomics refers to the investigation of microbiome diversity via all genes occuring in an environmental sample – as opposed to via one gene (usually 16S rRNA) alone. You can learn more in the linked review papers here and here if you wish.
Prof. Runstadler will address 16S rRNA approaches in lecture this week, and you can also refer to the Day 1 and Day 2 wiki introductions.
You can learn about UniFrac software at the linked paper or the direct site. It was used by the authors for some of their analyses.
Discussion Topics
Writing
As you read the paper by Koenig et al., consider not only its scientific content, but also the authors' writing style (perhaps not all on one read!). Sketch out answers to the questions below (right on the paper if you wish). Your answers will not be collected, but you may be called on in discussion to share your ideas.
- What functional elements does the abstract contain? As a whole, did the abstract make you want to read the paper?
- Now consider the Introduction section.
- What is the topic and/or function of each paragraph?
- How closely does this introduction conform to the suggested three-section structure described in the class scientific writing guidelines? Is there too much or too little information or emphasis on any particular topic?
- What purpose(s) do the citations serve?
- Now consider the Results section.
- What purpose do the sub-section titles serve? Which ones do so most effectively?
- Can you find one or more examples of paragraphs with effective introductory and concluding sentences, according to the description here?
- How about examples with ineffective opening and closing sentences? How might you improve these?
- Are there any parts in the Results that you think belong in the Discussion instead, according to the descriptions here and here ?
- Finally, consider the Discussion section.
- What is the topic and/or function of each paragraph?
- Is there too much or too little information or emphasis on any particular topic?
- Have the authors framed their writing to suggest future studies?
- What purpose(s) do the citations serve?
Content
You were previously assigned one of the topics below to present to and discuss with the rest of the class. We'll now break to listen to Atissa's talk about giving talks, then give you some time to revise the slide that you prepared, and finally go through the slides and topics one by one. Per slide, we'll first discuss scientific content, and then give you feedback about your slide design and presentation -- briefly and informally.
- Figure 1 – assigned to Yellow Team
- How is phylogenetic diversity (PD) defined? What general trend was observed? How do the authors interpret various exceptions to the trend?
- Figure 2 – assigned to Purple Team
- Skim the linked paper about UniFrac software, and refer to it to explain the basic idea of principal coordinates analysis to your peers. What additional information is learned in Fig 2 as compared to Fig 1?
- Figure 3 – assigned to Grey Team
- What changes in bacterial population do the authors observe over time and to what life events do they attribute them? Which timespan population would you expect to vary the most among different infants, and which would you expect to be most similar across different infants?
- Figure 4 – assigned to Orange Team
- Briefly, define what C-score and checkerboard analysis measure. How did the authors decide whether these values were higher, lower, or the same as might be expected? What primary conclusion do they draw?
- Figures S1 + S2 – assigned to Blue Team
- What general trends in SCFA concentrations and bacterial load did the authors find, with respect to time? How did the authors approach error measurement, and how reliable does each dataset appear to you?
- Figure 5 (focus on 5C) – assigned to Pink Team
- Make sure you understand (and can explain) how to read a correlation matrix before you begin. How do the authors interpret the correlations they find when looking at bacterial diversity and metabolite concentrations together?
- If you have time to look at 5A and 5B... do the figures appear to accurately reflect the text?
- Figure 6A + S4 – assigned to Red Team
- What is the difference between the approach the authors take in 6A and S4? How do they explain the differences in their findings by said methods?
- Figure 6B + Table S5 – assigned to Green Team
- Describe a few trends in gene abundance that the authors found. How easily can you discern these findings from the figure and table? How might you redesign the figure and/or table for easier use by the reader?
We’ll briefly discuss Figures S3 and Table S6 as a group.
For next time
Because of the holiday next week, this assignment is long. Be sure to start it earlier than the night before our next lab day. Parts 1 and 2 of this assignment are also due on Stellar.
- You will write up the work you do in Module 1 in a formal lab report. To help you pace your work, as well as give you feedback early on, you will be required to draft small portions of the report as homework assignments. For next time, you should write an early draft of your Materials and Methods: on DNA extraction and PCR. Be sure to read the Methods section guidelines at this link before you begin; doing so may save you some effort.
- Begin drafting the Introduction section of your report. See the specific assignment as well as general writing guidelines for suggestions about structure and content. Specifically, at a minimum you should hand in
- The first 4-5 sentences of your introduction -- these are crucial for capturing the reader's attention!
- The last paragraph of your introduction (excluding the preview of results).
- A high-level outline of what goes in between, plus a few notes!
- By high-level outline, I mean something approximating the opening and closing sentences for each paragraph. These can be written as phrases rather than complete sentences. The opening should establish the topic, and the closing should suggest a transition from one topic to the next. (A typical introduction is 3-5 paragraphs total.)
- By notes, I mean that below each topic sentence you should jot down a few notions about (a) relevant points that you already are familiar with and (b) topics that you know you need to research further.
- You do NOT have to combine the above two elements into some over-structured, Roman-numeral filled, "classic" outline. You will figure out how everything truly goes together as you get further along in the lab and in your writing.
- Note: You are welcome to hand in complete sentences for the entire introduction if you want to, but please also attach some kind of outline/notes that capture your thought process as you went about drafting it.
- The other major assignment you will complete during Module 1 is your primer design summary. While the information is fresh in your mind, turn your M1D1 notes into a table appropriate for submission. Eventually, you will write 1-2 paragraphs summarizing the motivation for your design challenge as well as explaining your design choices. For now, simply write 3-5 sentences summarizing the major differences between your primers and the original primers, and explaining how you expect the new primers will behave as a result of these differences.
Reagent list
- PfuUltra polymerase and buffer from Agilent
- dNTPs from Promega, original stock at 10 mM of each base
- Primers
- intermediate stock concentration is 5 μM for each primer (5 μM total), diluted from individual 100 μM long-term stocks
- F8-27 sequence: 5' AGAGTTTGATCCTGGCTCAG
- R1392-1407 sequence: 5' ACGGGCGGTGTGTACA