20.109(F14): Mod 1 Day 3 Agarose gel electrophoresis
Agarose gel electrophoresis
Introduction
Electrophoresis is a technique that separates large molecules by size using an applied electrical field and a sieving matrix. DNA, RNA and proteins are the molecules most often studied with this technique; agarose and acrylamide gels are the two most common sieves. The molecules to be separated enter the matrix through a well at one end and are pulled through the matrix when a current is applied across it. The larger molecules get entwined in the matrix and retarded; the smaller molecules wind through the matrix more easily and travel further from the well. Molecules of the same size and charge migrate the same distance from the well and collect into a band.

DNA and RNA are negatively charged molecules due to their phosphate backbone, and they naturally travel toward the positive charge at the far end of the gel. They are typically examined with agarose gels. Proteins are composed of amino acids that can be positively, negatively or uncharged. To give proteins a uniformly negative charge, they are coated with a detergent, SDS, prior to running them on a gel. Protein samples are also boiled to remove any secondary structure that might make two molecules of the same size migrate differently. Polyacrylamide is the matrix commonly used to separate proteins. These gels are typically run vertically while agarose gels are run horizontally but gravity has nothing to do with the separation.
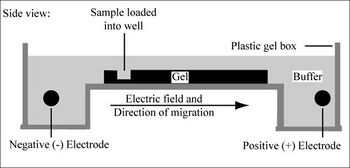
Today you will separate DNA fragments using an agarose matrix. Agarose is a polymer that comes from seaweed and if you’ve ever made Jell-O™, then you already have all the skills for pouring an agarose gel. To prepare these gels, agarose and buffer are microwaved until the agarose is melted. The molten agar is then poured into a horizontal casting tray, and a comb is added. Once the agar has solidified, the comb is removed, leaving wells into which the DNA sample can be loaded.
The distance a DNA fragment travels is inversely proportional to its length. Over time fragments of similar length accumulate into “bands” in the gel. Higher concentrations of agarose can be used to resolve smaller DNA fragments. The figure below (far right) shows the same DNA fragments resolved with three agarose concentrations. The 1000 base pair fragment is indicated in each.
- Agarose gel electrophoresis
-
Scanning EM image of agarose polymer
-
Agarose gel data analysis
-
Agarose gel raw data
Ethidium Bromide is a fluorescent dye that is commonly added to agarose gels. This dye intercalates between the bases of DNA, allowing DNA fragments to be located in the gel under UV light and photographed. The intensity of the band reflects the concentration of molecules that size, although there are upper and lower limits to the sensitivity of dyes. Because of its interaction with DNA, ethidium bromide is a powerful mutagen and will interact with the DNA in your body just as it does with any DNA on a gel. You should always handle all gels and gel equipment with nitrile gloves. Agarose gels with Ethidium Bromide must be disposed of as hazardous waste.
Today you will run your digested plasmid backbone and the digested PCR product on an agarose gel. You will then cut the relevant bands of DNA out of your gel and proceed to purify them from the agarose. Next time you will mix them in a ligation reaction.
Protocol
Part 1: Running your gel
You will use a 1% agarose gel (prepared by the teaching faculty), running seven samples as well as a reference lane of molecular weight markers (also called a DNA ladder).
- Add 20 μL of sterile water to the 5 μL PCR aliquots (not the PCR double digest) you stored after Mod1 Day2.
- Add 4 μL of 6X loading dye to the PCR aliquots, as well as to the pCX-NNX digested by XbaI and/or EcoRI, and the PCR product digested by XbaI/EcoRI.
- Loading dye contains bromophenol blue as a tracking dye to follow the progress of the electrophoresis (so you don’t run the smallest fragments off the end of your gel!) as well as glycerol to help the samples sink into the well.
- Xylene cyanol may also be used as a tracking dye, but does not migrate as far across the gel.
- Flick the eppendorf tubes to mix the contents, then quick spin them in the microfuge to bring the contents of the tubes to the bottom.
- Load the gel in the order shown in the table below.
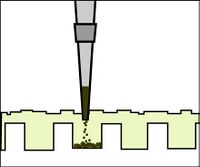
- To load your samples, draw the volume listed on the table below into the tip of your P200. Lower the tip below the surface of the buffer and directly over the well. You risk puncturing the bottom of the well if you lower the tip too far into the well itself (puncturing well = bad!). Expel your sample into the well. Do not release the pipet plunger until after you have removed the tip from the gel box (or you'll draw your sample back into the tip!).
- Once all the samples have been loaded, attach the gel box to the power supply and run the gel at 125 V for no more than 45 minutes.
- While the gels run, we will have our pre-lab lecture. Please also find some time to pre-weigh and label two eppendorf tubes before your gel is done!
- You will be shown how to photograph your gel and excise the relevant bands of DNA.
| Lane | Sample | Volume to load |
|---|---|---|
| 1^ | Uncut pCX-NNX^ | 10 μL^ |
| 2 | pCX-NNX XbaI | 5 μL |
| 3 | pCX-NNX EcoRI | 5 μL |
| 4 | pCX-NNX XbaI + EcoRI | 25 μL |
| 5 | 1Kb DNA Ladder | 20 μL |
| 6 | PCR Product XbaI + EcoRI | 25 μL |
| 7 | PCR Product Uncut | 25 μL |
| 8 | PCR no-template-control | 25 μL |
^Available from the teaching faculty
Part 2: Pre-lab lecture
Today we'll do our pre-lab lecture once everyone's gel is running!
Part 3: Isolating/Purifying DNA
To purify your DNA from the agarose, you will use another kit sold by Qiagen. Again the reagents have uninformative names and their contents are proprietary. Like the PCR Clean-up kit, the Agarose Purification kit requires binding the DNA to a spin-column with a silica-gel membrane, washing away salts and eluting the DNA from the membrane.
- Estimate the volume of your gel slices by weighing them.
- Add 3 volumes of QG for every 1 volume of agarose. (The maximum advised volume is 550 ul. Continue for now, but read step 5 carefully.)
- Incubate in the 50°C water bath for 10 minutes, until the agarose is completely dissolved. Every few minutes, you can remove your tubes from the 50°C heat to flick the contents. This agitation will help dissolve the agarose.
- Add 125 μL of isopropanol to each eppendorf tube.
- Get two QIAquick columns and two collection tubes from the teaching faculty. Label the spin-column (not the collection tubes!) either “bkb” or “frag” then pipet the appropriate dissolved agarose mixture to the top. Microfuge the column in the collection tube for 60 seconds. The maximum capacity of the QIAquick columns is 800 uL! If you have more than 800 uL in your mixture, you will need to repeat this step on the same column.
- Discard the flow-through in a temporary waste container (such as a 15 mL conical tube) and replace the spin-columns in their collection tubes. Add 750 ul of PE to the top of the column and spin as before.
- Discard the flow-through in your waste container and replace the spin-columns in their collection tubes. Add nothing to the top but spin for 60 seconds more to dry the membrane.
- Trim the cap off two new eppendorf tubes and label the sides with your team color, the date, and either “bkb” or “frag.” Place the spin-column in the correct trimmed eppendorf tube and add 30 ul of EB to the center of the membrane.
- Allow the columns to sit at room temperature for one minute and then spin as before. The material that collects in the bottom of the eppendorf tubes is your purified plasmid backbone or insert, ready to be ligated.
- At the end of the day, dump your waste tube into the chaotropic salt/ethanol waste in the fume hood, and save the tube for another day.
Part 4: Evaluate recovery
Ligations generally work best when there is a 1:4 molar ratio of backbone to insert. You will ligate your fragments next time, but before you do, run a small amount out of the purified products on a gel. This will allow you
- to check that you purified your DNA from the agarose
- to assess the quality of the DNA (that it’s not degraded or contaminated)
- to adjust the amount of backbone and fragment in your ligations for a ~1:4 ratio.
- Move 5 μL of each sample into an eppendorf tube labeled “purif bkb” or “purif frag.” Add 15 μL sterile water to each tube and 3 μl loading dye. Use a colored label to identify your team color and give the samples to the TA. The TA will run a 1% agarose gel for you and the result will be posted. Once it is posted, you may complete the FNT #2 to prepare for the ligation you will do in lab next time.
- Freeze the remainder of your purified DNAs at –20°C.
DONE!
For next week (Due M1D4)
Part 1 of this assignment is also due on Stellar.
- In addition to the methods section that you began last time, you will document the work you do during Module 1 in a formal written summary. To help you pace your work, as well as give you feedback early on, you will be required to draft small portions of the summary as homework assignments. For next time, you should prepare a page on today's experimental outcomes.
- Begin by reading, under Logistics, Guidelines on Formatting and Length. Next, skim the Content Guidelines, paying particular attention to those relevant for M1D3.
- You should present both the purification gel and the recovery gel, each in a well-labeled figure with an appropriate figure caption. Be sure to follow our written guidelines as well as the suggestions presented by your writing instructor during class.
- Below the figures, interpret the outcomes in concise language. Consider reviewing slides 30 and 31 in the Writing Lab talk titled EBICS REU Poster Best Practices DMC SWO 1 to learn how to turn bulky sentences into succinct bullets. It's also okay to use complete sentences when they more clearly make your point.
- Please see this example for a suggested layout.
- To prepare for our next lab session, calculate the amounts of DNA you will use in the ligation by following the steps below.
- Separately calculate the concentration of backbone and of insert based on the recovery gel posted on today's Talk page. Refer to the NEB marker definitions to estimate the ng of DNA in each lane, and refer to your notebook/protocol for the relevant volume basis. Note that the ng listed are for 10 μL of ladder.
- You may convert the mass concentration to a molar concentration, using the fact that a typical DNA base is 500 g/mol. This conversion will mostly cancel out between the insert and the backbone, except for the difference in number of bases. Feel free to either omit steps that will cancel if you are comfortable doing so, or to keep them if you follow the lab math better that way.
- Ideally, you will use 50-100 ng of backbone in the upcoming ligation. Referring to the mass concentration, what volume of DNA will this amount require?
- Ideally, you will use a 4:1 molar ratio of insert to backbone. Referring to the molar concentrations, how much insert do you need per μL of backbone?
- A 15 μL scale ligation should not include more than 13.5 μL of DNA. If your backbone and insert volumes total to greater than this amount, you must (1) scale down both DNA amounts, using less than 50 ng backbone and/or (2) stray from the ideal 4:1 molar ratio. You may ask the teaching faculty for advice during class if you are unsure what choice is best, but make and submit an initial ligation plan for now.
Reagents list
- NEB Loading Dye (link)
- 2.5% Ficoll®-400
- 11mM EDTA
- 3.3mM Tris-HCl
- 0.017% SDS
- 0.015% bromophenol blue
- pH 8.0@25°C
- NEB 1 kb ladder (link)
- 1% agarose gel in 1X TAE
- 1X TAE
- 40 mM Tris
- 20 mM Acetic Acid
- 1 mM EDTA, pH 8.3
- QIAquick gel extraction kit
- isopropanol
Navigation Links
Next Day: Mod 1 Day 4: Ligation & Transformation Previous Day: Mod 1 Day 2: Clean and cut DNA