Biomod/2011/TeamJapan/Sendai/Notes/Stick
<html>
<style rel="stylesheet" type="text/css">
.clear {clear:both;}
#verticalmenu {
/*this .CSS is inspired by http://www.javascriptkit.com/ */ font-family: "Comic Sans MS" , "Brush Script MT",serif, sans-serif, monospace, cursive, fantasy; list-style:none;}
- verticalmenu a:hover {
color: #aa1d1d; /* color when the click is over the main menu text-transform: uppercase; font-size: 10px; */
}
a:visited { color:#00a5ea; text-decoration: none }
.glossymenu, .glossymenu li ul{ list-style-type: none; margin: 0; padding: 0; width: 250px; /*WIDTH OF MAIN MENU ITEMS*/ border: 1px solid black; list-style:none; }
.glossymenu li{ position: relative; }
.glossymenu li a{ background: white url(http://openwetware.org/images/a/a7/Glossyback2.jpg) repeat-x bottom left; font: bold 17px Verdana, Helvetica, sans-serif; color: white; display: block; width: auto; padding: 10px 0; padding-left: 10px; text-decoration: none; }
.glossymenu li ul{ /*SUB MENU STYLE*/ position: absolute; width: 200px; /*WIDTH OF SUB MENU ITEMS*/
left: 0; top: 0; display: none; }
.glossymenu li ul li{ float: left; }
.glossymenu li ul a{ width: 190px; /*WIDTH OF SUB MENU ITEMS - 10px padding-left for A elements */ }
.glossymenu .arrowdiv{ position: absolute; right: 2px; background: transparent url(http://openwetware.org/images/6/69/Arrow.gif) no-repeat center right; }
.glossymenu li a:visited, .glossymenu li a:active{ color: white; }
.glossymenu li a:hover{ background-image: url(http://openwetware.org/images/5/50/Glossyback.jpg); }
/* Holly Hack for IE \*/
- html .glossymenu li { float: left; height: 1%; }
- html .glossymenu li a { height: 1%; }
/* End */
</style>
<script type="text/javascript">
/***********************************************
- CSS Vertical List Menu- by JavaScript Kit (www.javascriptkit.com)
- Menu interface credits: http://www.dynamicdrive.com/style/csslibrary/item/glossy-vertical-menu/
- This notice must stay intact for usage
- Visit JavaScript Kit at http://www.javascriptkit.com/ for this script and 100s more
- /
var menuids=new Array("verticalmenu") //Enter id(s) of UL menus, separated by commas var submenuoffset=-2 //Offset of submenus from main menu. Default is -2 pixels.
function createcssmenu(){ for (var i=0; i<menuids.length; i++){
var ultags=document.getElementById(menuids[i]).getElementsByTagName("ul")
for (var t=0; t<ultags.length; t++){
var spanref=document.createElement("span")
spanref.className="arrowdiv" spanref.innerHTML=" " ultags[t].parentNode.getElementsByTagName("a")[0].appendChild(spanref)
ultags[t].parentNode.onmouseover=function(){
this.getElementsByTagName("ul")[0].style.left=this.parentNode.offsetWidth+submenuoffset+"px"
this.getElementsByTagName("ul")[0].style.display="block"
}
ultags[t].parentNode.onmouseout=function(){
this.getElementsByTagName("ul")[0].style.display="none"
}
}
}
} if (window.addEventListener) window.addEventListener("load", createcssmenu, false) else if (window.attachEvent) window.attachEvent("onload", createcssmenu) </script>
|
<img src="http://openwetware.org/images/0/07/3D_sendai.png" width="650"> |
</html> <html> <head> <style type="text/css">
- content {padding-left: 10px;width: 970px;}}
h3 {font-decoration: none;} h1.firstHeading {display: none; } </style> </head> </html>
<html> <style rel="stylesheet" type="text/css"> /*このスタイルシートの著作権はテンプレート工房TAKEにあります*/ /*ページのレイアウト用css*/
body{ background:#F5F5DC; /*壁色と壁紙設定*/ background-repeat:repeat;/*繰り返さない場合はno-repeatに変更*/ font:"メイリオ", "MS Pゴシック", Osaka, "ヒラギノ角ゴ Pro W3"; color: #333333; margin:0px; padding:0px; }
- contents{
width:900px;
margin:0 auto; background-color: #FFFFFF ;/*コンテンツ内の背景(サイズをぴったりにすること)*/ background-repeat:repeat-y; /*縦に繰り返し*/ border:solid 1px #666666;/*サイトに枠を付ける設定,色の変更可*/
position:relative;
font-size:80%;
}
/*ヘッダー部分の設定*/
- header{
background-image:url(http://openwetware.org/images/c/c4/TeamSendai-logo3.png) ;/*ヘーダー*/ background-repeat:repeat-x; /*縦に繰り返し*/ background-position:top right; height:140px; /*ヘーダーの高さ*/ }
- header p {
font-size: 26px;
color:#333333;
padding-top: 15px; padding-left: 20px; }
/*上部メニューボタンの設定*/
- navbar{
background-color:#FFFFFF;
width: 100%;
height:40px;
position:margine;
top:100px;
left:0px;
border-top:solid 1px #FFFFFF;
border-bottom:solid 1px #FFFFFF;
}
- navbar ul{
margin:0;
padding:0; list-style-type:none; font-family:Arial, Helvetica, sans-serif; font-size: 12px; line-height:40px; letter-spacing:2px; }
- navbar li{
background-color:#000099; /*上部メニューのボタンの背景*/
float:left; width:146px; /*メニューボタンの幅*/ text-align:center; padding:0; border-right:solid 1px #ffffff; }
- navbar ul a:hover{
background-color:#0033cc; /*メニューボタンにカーソルが来た時に背景*/
width:146px; /*メニューボタンの幅*/ }
- navbar a{
color:#ffffFF;/*メニューボタンの文字の色*/
display:block; }
- navbar a:hover{
color:#999999; /*メニューの文字がカーソルが来た時、この色に変わる*/ }
/*サイドメニューの設定*/
- side{
background-color:#ddffff;
width:220px;/*サイドの幅(変更するときはコンテンツ背景も変更すること)*/
position:margin; top:600px;/*上からの位置*/ left:12px; }
- side h3 {
font-size: 90%; border: double 3px #FFFFFF; color:#ffffff; text-align: center; background-color:#999999;
width:190px;
line-height: 30px; margin-top: 10px; margin-left: 5px; margin-bottom: 5px; }
- side h3 a {
color:#ffffff;
font-weight:normal; }
- side ul{
font-size:100%;
line-height:220%; /*サイドの文字と文字の行間設定*/ background-color: #ddffff; margin:0px; padding-left:10px; }
- side ul a:hover {
width:180px;
background-color: #99ffff; /*サイドのカーソルオーバー時の背景色*/ color: #999999; /*サイドのカーソルオーバー時の文字色*/ }
- side ul{
list-style-type:none;
padding-left:2px; }
- side li{
padding-left:15px; /*文字の左端からの位置*/ }
- side li a{
color:#333333;/*サイドの文字色*/
width:180px;
display:block;
}
- side .ad_list li{
background-image:none;
padding-left:0; }
/*右側メイン部分の設定*/
- main{
width:630px;
padding-top:15px;
margin-left:240px;
}
/*下部のフッター部分の設定*/
address{
font-size:80%;
font-style:normal;
text-align:center;
padding-top:5px;
}
address{
background-color:#000066;
color:#ffffff;
width:882px;
padding-bottom:10px; border:none; } address a{
color:#ff9999;
}
/*文字の設定*/ h1{ font-size:60%; letter-spacing: 2px; padding-left:10px; margin: 0px; }
h1 a{
color:#FFFFFF;
font-weight:normal; }
h2{
font-size:140%;
border-left: 10px solid #000066;
border-bottom:solid 1px #000099;/*文字の下に線を入れる設定*/
width:900px;
padding-left: 5px; color:#333333; margin-top: 15px; margin-bottom: 5px; }
h3{
font-size:120%;
border: solid 1px #111111;
color:#ffffff;
background-color:#4682B4 ; line-height: 30px; padding-left:10px; margin-top: 10px; margin-bottom: 1px; }
p{
font-size:90%;/*全体の文字サイズ*/
line-height:150%;/*全体で使う、文字と文字の行間*/
margin-left:5px;
}
p img{
float:left;
margin-top:5px; /*写真の左にスペースを空ける*/
margin-left:5px; /*写真の左にスペースを空ける*/
margin-right:10px; /:写真と文字の間隔*/ }
/*リンク文字の設定*/
a{
color:#000099;
text-decoration:none;
} a:hover { color: #FF0000;/*リンクの文字の上にマウスが来た時この色に変わる*/ text-decoration: none; }
- purple{
font-size:120%;
border: solid 1px #111111;
color:#ffffff;
background-color:#9370DB; line-height: 40px; padding-left:10px; margin-top: 10px; margin-bottom: 1px; }
h5{
font-size:120%;
border: solid 1px #111111;
color:#ffffff;
background-color:#FFA500; line-height: 30px; padding-left:10px; margin-top: 10px; margin-bottom: 1px; }
h6{
font-size:120%;
border: solid 1px #111111;
color:#ffffff;
background-color:#006400; line-height: 30px; padding-left:10px; margin-top: 10px; margin-bottom: 1px; }
- red{
font-size:120%;
border: solid 1px #111111;
color:#ffffff;
background-color:#DC143C; line-height: 40px; padding-left:10px; margin-top: 10px; margin-bottom: 1px; }
- blue{
font-size:120%;
border: solid 1px #111111;
color:#ffffff;
background-color:#191970; line-height: 40px; padding-left:10px; margin-top: 10px; margin-bottom: 1px; }
</style> </html>
Robot Design
This design is a very simple one, where it is combined only four DNA double strands.The sticky motif robot was our plan B in case we failed to produce the triangular prism robot.In addition the legs design include both kind of robots for saving time and costs. It assumes that how to move is the same as a triangular prism.
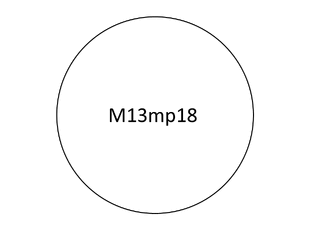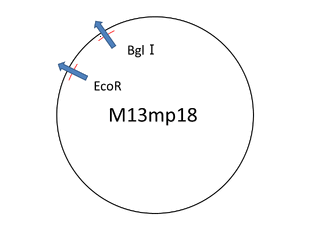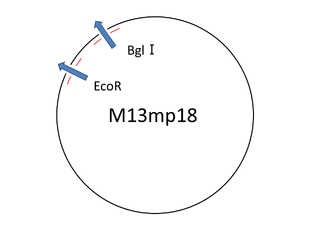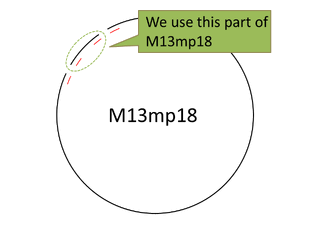
For producing sticky motif we used the viral M13mp18 DNA single strand (M13) as scaffold. But we only used 469 bases of 7,249 bases , then having a leftover. In this situation we thought about cutting the part of M13 that we needed. Our first attempt was to extract the necessary part of M13 to reproduce M13 with polymerase chain reaction. But we failed. So we changed our method for cutting the M13 with restriction enzyme(bglⅠ,EcoR).
 |
 |
Experiment
- Electrophoresis
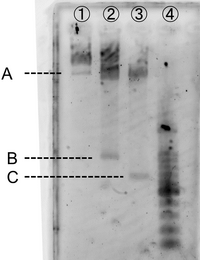
gel:1.0% agarose gel
buffer:1×TAE
For producing the sticky motif robot body, firstly we used whole M13mp18 DNA single strand (M13) as scaffold. However, 469 bases of 7,249 bases were used in our design. Thus, no used region of M13 was cut with restriction enzyme (Bgl I and EcoR). The reason why the band of C is smaller amount than the band of D is that the samples was purified by QIAGEN PCR purification kit, by which longer DNA (>5000bp) is not effectively purified, before loading gels.
Protocol
- Cut M13
To cut m13, specific part of m13 sequence form a double helix, secondly react by enzyme, finally clean up of discarded enzyme and pick out only M13.The process is shown below.
1.Form a double helix
Mix 5μL M13(84nM), 3μL DNA that specific part of m13 sequence form a double helix(5μM), 2μL 10×H buffer, and 11μL mQ.
Set in PCR, the condition is 3min at 95°C, 3min at 65°C, for the duration of enzyme reaction, let the sample at 37°C.
2.Cutting M13 by enzyme reaction
Add 0.5μL BglⅠ and EcoR in the sample, let the sample stand for 1 hours at 37°C.
3.Clean up of enzyme
This protocol is [here]
Conclusion
Since there was no time in this time, time was not able to be spared for sticy motif. Therefore, it did not go by electrophoresis to the check of structure. Structure is so small that we can not observed sticky motif under the AFM. So we wanted to check structure by whether sticy motif and the field which attached C-leg are mixed, and structure is attached to the position of the start, but a structure did not ride on the field, so a plan did not progress.
DNA sequence
ACCCGTCGGATTCTCCGTGG
GAACAAACGTTTGTTAATTTTTT
CGTATCGGCCTTTCGCTGACATTCAGGC
TAAATGTCAGCTCAAATTCGCGTTAAATTT
GATAGGTTACGTCAACAT
CTCCAGCCAGCTTTCCCTGCCAGTTTGAGGAAGATTGTAATCAGAA
TGGCCTTCCTGTAGCCAGCTTTCATTGGGAACGCCA
CGTAACCGTGCATGGCCCATTCGCTATAAGCA
TCAAAAATAATTCGCGTC
AATATTTAAATTGATAGTGTAGATGGGCGCAT
GAAACCAGCTGTTGGGAAG
TTGTTAAATGAGCGAGTAACA
AACCATAAACAAAAACAGGGGAATGG
TGCGCAAGCAAAGCGACCGCTTCTGGTGCCG
AAGCCCCGTTAATATGCGGATTGACCGTACGACGAC
GGCGATCGGTGCGGGCCTCCAGGAAGATCGCA