User talk:Jacqueline Nkuebe
Assignments
Final Thoughts
This semester I've been involved in: 1) Doing literature searches to try to come up with a heritable trait with little environmental influence: looked into LDL cholesterol, SNPs implicated in height, BMI and ultimately fell upon eye color with the help of Ridhi and Anu
2) Meeting almost weekly with the math modeling group
3) Contacting various scientists (Fan Liu, Amy Carmargo, and Bruce Birren) to gain insight into the complexities of creating an epistatic tool in Trait-o-matic): tackling issues of absolute chromosomal location, access to data on HapMap
4) Along with Ridhi, the two of us wrote the proposal to the Rotterdam Management Team
5) Attempts at finding a dataset in HapMap
6) Working diligently with Anu and Ridhi to think of ways of improving the usability of Trait-o-Matic.
For more details, see the Biowiki page.
SEE BIOLOGY GROUP Page for Further Correspondence
OCTOBER 29TH Thoughts on Current Project and Projected Contributions
My major concerns going into the class project were: 1. A lack of programming experience. and 2. Wanting to make sure that I was contributing equally to the project (even if that's in a non-programming role).
I'm most comfortable with: 1. Thinking of unique ways to organize data 2. Interfacing 3. Data mining
I think that the best way for us to all gain the most of the experience may be to couple people with similar interests (2 to 3) to work on subprojects, where in at least one person feels most comfortable with programming and the other(s) feels most comfortable with their knowledge of molecular bio/biochem/metabolics, ect. so that we can share knowledge.
OCTOBER 20TH Additional Research
In thinking about the most recently suggested class project, SNPCupid, I decided to check out many of the current genetic risk testing services available so that we might consider either augmenting or using similar services in our own web tool. Though services like deCODEme are for individual use outside of the pre-natal, could be eugenic sector, much of the information they provide is quite useful. I think that for SNPCupid, in addition to using genetic analysis to predict the likelihood of disease predispositions in offspring, we might consider adding a few things:
1) I think that it would be great in analyzing particular disease hits to have some sort of analysis that gives the number of geneitc variants known to increase risk for a particular disease, and to give some stats about the percentage of people tested who had similar/the same variant. It might go something like this: I'll use psoriasis as an example. "There are eight genetic variants known to increase the risk of developing psoriasis; located on chromosomes 1, 5, 6, and 12. Of these the variant in the HLA-C gene contributes by far the strongest effect to the risk of developing psoriasis in most if not all populations tested."
2) I think that it would also be useful to provide users with services for prevention and treatment. We could consider making stats on the particular disease predispositions for their future child made available maybe based on race, gender, and take a look at current mortality or remission rates. We could also provide links to genetic counseling services, or even recommend clinicians for our clients. With regard to ethnic stats for psoriasis, SNPedia provides the following information:
psoriasis Europeans
* rs20541(G) 1.27 * rs610604(G) 1.19 * rs2066808(A) 1.34 * rs2201841(G) 1.13 * rs4112788(G) 1.41
Chinese
* rs1265159(A) 22.62 Proxy SNP for rs1265181 * rs3213094(C) 1.28 * rs4085613(G) 1.32
3) This maybe kind of "out there" but we could also have a network wherein our clients could connect with the parents of children who have various genetic diseases.
4) We could also provide services for mutation/variant conformation. Since we may not request to know what services our client used for their genome sequencing, it might be useful (if we get a variant "hit") to provide them with tools to be able to verify the call we get...or we could do it ourselves.
OCTOBER 13TH AND 20TH ASSIGNMENTS (Considering the other two gene databases)
GeneCards seems like it would be useful in gathering information about the PharmaGKB drug compound relationships in addition to analyzing some biochemical/molecular bio consequences of SNPs and variants for CYP450 genes. Looking through the GeneCards database, I found the disorders and mutations subsection may act to complicate the pharmacogenomic dosage tool that we're interested in. Many allelic variations in CYP450 genes not only affect drug metabolism but contribute to disease profiles. I wonder how complicated it would be to prescribe an appropriate dosage of a drug if someone is also dealing with a pre-existing condition. We may run into the problem of how to recommend a correct dosage of a particular agent in such a way that it is both efficacious but does not cause harmful affects due to drug-combinations for a pre-existing disease.
With regard to my concern about people with pre-existing diseases and their use of the drug dosage tool: I took a look at one CYP450 gene, CYP1A1, and found that there are several mutations associated with CYP1A1 that may affect not only drug metabolism, but cancers. NovoSeek disease relationships with CYP1A1 also suggested that mutations in this gene are responsible for breast cancer and squamous cell carcinoma. CYP1A1.
Emerging Ideas: Since I've missed the last few lectures, I was surprised to see this emerging proposal for SNPCupid that's been proposed. I saw Joe's comments on the proposal and I see interesting potential in the project. I also understand that this proposal might be an easier way to get everyone to participate via our use of Phyton. I probably missed the discussion as to why the Pharmacogenomic tool may not work, but I actually find the Drug Dosage proposal more interesting. I'm sure that this is something that we will discuss in class.
OCTOBER 1st
CYP450 isoforms and drug metabolism: Thought-experiments on how to use current databases to realize Filip's Idea for a Pharmacogenomic Drug Dosage Tool
Known Information: OMIM has a nice description with primary literature accompaniment describing CYP2C9, one of the major drug-metabolizing CYP450 isoforms. Moreover, NCBI, always on the forefront of bioinformatics research, has developed database repositories for both SNP (dbSNP) and microarray (GEO) data. Access to these data and information resource can be used not only to help determine drug response but to study disease susceptibility and conduct basic research in population genetics. Another really cool documentation on OMIM was work done by Wood in 2001, in which he describes the difference in pharmacogenetics between different races. It might be interesting to see how allelic variations for particular CP450 genes may depend on race. This, of course, will depend a lot on the availability of more genomes.
Information to be Researched: SNPedia has a fantastic list of SNPs in CYP2C9 but many of the phenotypes for particular mutations are undefined. This suggests some practical limitations to understanding how a particular SNP may affect drug metabolism. We should consider raking through literature to see if there are important environmental dispositions that affect drug metabolism. Some of the major ones include nutrition, alcohol/smoking, drug-drug interactions and liver function and size. It's also important to consider epistatic effects, as well.
Strategy: In addition to some of the above mentioned information, see Filip's Diagram.
Future: SNP screenings will benefit drug development and testing because pharmaceutical companies could exclude from clinical trials those people whose pharmacogenomic screening would show that the drug being tested would be harmful or ineffective for them. Excluding these people will increase the chance that a drug will show itself useful to a particular population group and will thus increase the chance that the same drug will make it into the marketplace. Also, talking to Brett, I think it might be interesting to include observational data. So instead of just telling a user " you should take this dose" we could say " because you are X, we think you should take this dose." Y percent of users of your type had this result."
SEPTEMBER 29th: Familiarize yourself with OMIM, GeneTests, and SNPedia and note on your talk page at least one big concept for Human2.0 and at least one small step we could take toward achieving it. Please use hypertext to indicate resources, references and/or collaborative work within our class (if any).
For many years the majority of concentrated effort in understanding human disease has gone into looking for a gene or set of genes that have acquired mutations spontaneously and given rise to rogue protein functionality. The underdog in the hierarchy of 'cool' biology has been the intron, once called 'junk DNA.' Recently, however, it has been demonstrated that introns can play a significant role in gene regulation. Similarly, 5' UTRs usually contain common regulators of expression: motifs, boxes, response or binding elements. 3' UTRs are also involved in gene expression although they do not contain well-known transcription control sites. 3' UTR sequences (or adenylation control elements) can control the nuclear export, polyadenylation status, subcellular targeting, rates of translation and degradation of mRNA. With regard to Human 2.0 I propose a diagnostic project in which we develop a web tool that analyzes mutations in UTRs and introns and their effect on downstream (or upstream) mRNA regulation and subsequent protein expression. Phenotyping could also be included. Relevance? One example, p53, the "guardian of the genome," has a well known and frequent intronic mutation implicated in familial breast cancer: p53 intron 2 mutation
Just to recap:
1. Diagnostics: A web tool that analyzes mutations in UTRs and introns and their affect on mRNA and subsequent protein expression.
- One step in reaching this goal is to check the literature to see if any such experimentation exists. We could collect data from the current literature and allow scientists and clinicians to add to it.
2. Random thought--Treatment: Cellularly Engineered Immune Cells that are resistant to HIV-1
- Since we neither have a grant from NIH nor a lot of people in this course willing to dedicate years in a laboratory, this idea is not feasible. I thought I would throw it out since I think it's kind of cool.
- This past spring, Nature documented one of the first phase I trial engineered immune cell technologies for fighting prostate cancer: Very Cool Cellular Engineering
SEPTEMBER 24th: List comprehensions saved my life, especially in parts 3 and 4.
p53_seg="cggagcagctcactattcacccgatgagaggggaggagagagagagaaaatgtcctttaggccggtt
cctcttacttggcagagggaggctgctattctccgcctgcatttctttttctggattacttagttatggcctttgc
aaaggcaggggtatttgttttgatgcaaacctcaatccctccccttctttgaatggtgtgccccaccccccgggtc
gcctgcaacctaggcggacgctaccatggcgtagacagggagggaaagaagtgtgcagaaggcaagcccggaggca
ctttcaagaatgagcatatctcatcttcccggagaaaaaaaaaaaagaatggtacgtctgagaatgaaattttgaa
agagtgcaatgatgggtcgtttgataatttgtcgggaaaaacaatctacctgttatctagctttgggctaggccat
tccagttccagacgcaggctgaacgtcgtgaagcggaaggggcgggcccgcaggcgtccgtgtggtcctccgtgca
gccctcggcccgagccgttcttcctggtaggaggcggaactcgaattcatttctcccgctgccccatctcttagct
cgcggttgtttcattccgcagtttcttcccatgcacctgccgcgtaccggccactttgtgccgtacttacgtcatc
tttttcctaaatcgaggtggcatttacacacagcgccagtgcacacagcaagtgcacaggaagatgagttttggcc
cctaaccgctccgtgatgcctaccaagtcacagacccttttcatcgtcccagaaacgtttcatcacgtctcttccc
agtcgattcccgaccccacctttattttgatctccataaccattttgcctgttggagaacttcatatagaatggaa
tcaggatgggcgctgtggctcacgcctgcactttggctcacgcctgcactttgggaggccgaggcgggcggattac
ttgaggataggagttccagaccagcgtggccaacgtggtg"
standard = { 'ttt': 'F', 'tct': 'S', 'tat': 'Y', 'tgt': 'C',
'ttc': 'F', 'tcc': 'S', 'tac': 'Y', 'tgc': 'C',
'tta': 'L', 'tca': 'S', 'taa': '*', 'tga': '*',
'ttg': 'L', 'tcg': 'S', 'tag': '*', 'tgg': 'W',
'ctt': 'L', 'cct': 'P', 'cat': 'H', 'cgt': 'R',
'ctc': 'L', 'ccc': 'P', 'cac': 'H', 'cgc': 'R',
'cta': 'L', 'cca': 'P', 'caa': 'Q', 'cga': 'R',
'ctg': 'L', 'ccg': 'P', 'cag': 'Q', 'cgg': 'R',
'att': 'I', 'act': 'T', 'aat': 'N', 'agt': 'S',
'atc': 'I', 'acc': 'T', 'aac': 'N', 'agc': 'S',
'ata': 'I', 'aca': 'T', 'aaa': 'K', 'aga': 'R',
'atg': 'M', 'acg': 'T', 'aag': 'K', 'agg': 'R',
'gtt': 'V', 'gct': 'A', 'gat': 'D', 'ggt': 'G',
'gtc': 'V', 'gcc': 'A', 'gac': 'D', 'ggc': 'G',
'gta': 'V', 'gca': 'A', 'gaa': 'E', 'gga': 'G',
'gtg': 'V', 'gcg': 'A', 'gag': 'E', 'ggg': 'G'}
Part 1.
x= p53_seg.count("c")
y= p53_seg.count("g")
print ((float(x+y))/len(p53_seg))*100
Part 2.
new=p53_seg.replace("c", "x")
new2=new.replace("a", "y")
new3=new2.replace ("g", "c")
new4=new3.replace("t", "a")
new5=new4.replace("x", "g")
new6=new5.replace("y", "t")
print new6[::-1]
Part 3.
codon1=[p53_seg[3*s:(3*s)+ 3] for s in range(0, (len(p53_seg))/3)]
trans1=[standard.get(c) for c in codon1]
print trans1
codon2=[p53_seg[(3*s)+1:(3*s)+4] for s in range(0, (len(p53_seg)-1)/3)]
trans2=[standard.get(c) for c in codon2]
print trans2
codon3=[p53_seg[(3*s)+2:(3*s)+5] for s in range(0, (len(p53_seg)-2)/3)]
trans3=[standard.get(c) for c in codon3]
print trans3
rev=p53_seg[::-1]
codonminus1=[rev[3*s:3*s+3] for s in range(0, (len(p53_seg))/3)]
transminus1=[standard.get(c) for c in codonminus1]
print transminus1
rev=p53_seg[::-1]
codonminus2=[rev[3*s+1:3*s+4] for s in range(0, (len(p53_seg)-1)/3)]
transminus2=[standard.get(c) for c in codonminus2]
print transminus2
rev=p53_seg[::-1]
codonminus3=[rev[3*s+2:3*s+5] for s in range(0, (len(p53_seg)-2)/3)]
transminus3=[standard.get(c) for c in codonminus3]
print transminus3
Part 4.
mutations= {'c': 'g', 'a': 'g', 't': 'g', 'g': 'g'}
p53_seg_mut= [mutations.get(p53_seg[i]) for i in range(0, len(p53_seg), 100)]
mutation1 = list(p53_seg)
for i in range(0, len(p53_seg_mut)):
mutation1[i*100] = p53_seg_mut[i]
codon1=["".join(mutation1[(3*s):(3*s)+3]) for s in range(0, len((mutation1))/3)]
print codon1
trans1=[standard.get(c) for c in codon1]
print trans1
IDLE: ['G', 'S', 'S', 'S', 'L', 'F', 'T', 'R', '*', 'E', 'G', 'R', 'R', 'E', 'R', 'E', 'N', 'V', 'L', '*', 'A', 'G', 'S', 'S', 'Y', 'L', 'A', 'E', 'G', 'G', 'C', 'Y', 'S', 'R', 'P', 'A', 'F', 'L', 'F', 'L', 'D', 'Y', 'L', 'V', 'M', 'A', 'F', 'A', 'K', 'A', 'G', 'V', 'F', 'V', 'L', 'M', 'Q', 'T', 'S', 'I', 'P', 'P', 'L', 'L', '*', 'M', 'V', 'C', 'P', 'T', 'P', 'R', 'V', 'A', 'C', 'N', 'L', 'G', 'G', 'R', 'Y', 'H', 'G', 'V', 'D', 'R', 'E', 'G', 'K', 'K', 'C', 'A', 'E', 'G', 'K', 'P', 'G', 'G', 'T', 'F', 'E', 'N', 'E', 'H', 'I', 'S', 'S', 'S', 'R', 'R', 'K', 'K', 'K', 'K', 'N', 'G', 'T', 'S', 'E', 'N', 'E', 'I', 'L', 'K', 'E', 'C', 'N', 'D', 'G', 'S', 'F', 'D', 'N', 'W', 'S', 'G', 'K', 'T', 'I', 'Y', 'L', 'L', 'S', 'S', 'F', 'G', 'L', 'G', 'H', 'S', 'S', 'S', 'R', 'R', 'R', 'L', 'N', 'V', 'V', 'K', 'R', 'K', 'G', 'R', 'A', 'R', 'R', 'R', 'P', 'C', 'G', 'P', 'P', 'C', 'S', 'P', 'R', 'P', 'E', 'P', 'V', 'L', 'P', 'G', 'R', 'R', 'R', 'N', 'S', 'N', 'S', 'F', 'L', 'P', 'L', 'P', 'H', 'L', 'L', 'A', 'G', 'G', 'C', 'F', 'I', 'P', 'Q', 'F', 'L', 'P', 'M', 'H', 'L', 'P', 'R', 'T', 'G', 'H', 'F', 'V', 'P', 'Y', 'L', 'R', 'H', 'L', 'F', 'P', 'K', 'S', 'R', 'W', 'H', '*', 'H', 'T', 'A', 'P', 'V', 'H', 'T', 'A', 'S', 'A', 'Q', 'E', 'D', 'E', 'F', 'W', 'P', 'L', 'T', 'A', 'P', '*', 'C', 'L', 'P', 'S', 'H', 'R', 'P', 'F', 'S', 'S', 'S', 'Q', 'K', 'R', 'F', 'I', 'T', 'S', 'L', 'P', 'S', 'R', 'F', 'P', 'T', 'P', 'P', 'L', 'F', '*', 'S', 'P', '*', 'P', 'F', 'C', 'L', 'L', 'E', 'N', 'F', 'I', '*', 'N', 'G', 'I', 'R', 'M', 'G', 'A', 'V', 'A', 'H', 'A', 'C', 'T', 'L', 'A', 'H', 'A', 'C', 'T', 'L', 'G', 'G', 'R', 'G', 'G', 'R', 'I', 'T', '*', 'G', '*', 'E', 'F', 'Q', 'S', 'S', 'V', 'A', 'N', 'V', 'V']
mutations= {'c': 'a', 'a': 'a', 't': 'a', 'g': 'a'}
p53_seg_mut= [mutations.get(p53_seg[i]) for i in range(0, len(p53_seg), 100)]
mutation2 = list(p53_seg)
for i in range(0, len(p53_seg_mut)):
mutation2[i*100] = p53_seg_mut[i]
codon2=["".join(mutation2[(3*s):(3*s)+3]) for s in range(0, len((mutation2))/3)]
print codon2
trans2=[standard.get(c) for c in codon2]
print trans2
['R', 'S', 'S', 'S', 'L', 'F', 'T', 'R', '*', 'E', 'G', 'R', 'R', 'E', 'R', 'E', 'N', 'V', 'L', '*', 'A', 'G', 'S', 'S', 'Y', 'L', 'A', 'E', 'G', 'G', 'C', 'Y', 'S', 'Q', 'P', 'A', 'F', 'L', 'F', 'L', 'D', 'Y', 'L', 'V', 'M', 'A', 'F', 'A', 'K', 'A', 'G', 'V', 'F', 'V', 'L', 'M', 'Q', 'T', 'S', 'I', 'P', 'P', 'L', 'L', '*', 'M', 'V', 'C', 'P', 'T', 'P', 'R', 'V', 'A', 'C', 'N', 'L', 'G', 'G', 'R', 'Y', 'H', 'G', 'V', 'D', 'R', 'E', 'G', 'K', 'K', 'C', 'A', 'E', 'G', 'K', 'P', 'G', 'G', 'T', 'F', 'K', 'N', 'E', 'H', 'I', 'S', 'S', 'S', 'R', 'R', 'K', 'K', 'K', 'K', 'N', 'G', 'T', 'S', 'E', 'N', 'E', 'I', 'L', 'K', 'E', 'C', 'N', 'D', 'G', 'S', 'F', 'D', 'N', '*', 'S', 'G', 'K', 'T', 'I', 'Y', 'L', 'L', 'S', 'S', 'F', 'G', 'L', 'G', 'H', 'S', 'S', 'S', 'R', 'R', 'R', 'L', 'N', 'V', 'V', 'K', 'R', 'K', 'G', 'R', 'A', 'R', 'R', 'R', 'P', 'C', 'G', 'P', 'P', 'C', 'S', 'P', 'R', 'P', 'E', 'P', 'V', 'L', 'P', 'G', 'R', 'R', 'R', 'N', 'S', 'N', 'S', 'F', 'L', 'P', 'L', 'P', 'H', 'L', 'L', 'A', 'S', 'G', 'C', 'F', 'I', 'P', 'Q', 'F', 'L', 'P', 'M', 'H', 'L', 'P', 'R', 'T', 'G', 'H', 'F', 'V', 'P', 'Y', 'L', 'R', 'H', 'L', 'F', 'P', 'K', 'S', 'R', 'W', 'H', '*', 'H', 'T', 'A', 'P', 'V', 'H', 'T', 'A', 'S', 'A', 'Q', 'E', 'D', 'E', 'F', 'W', 'P', 'L', 'T', 'A', 'P', '*', 'C', 'L', 'P', 'S', 'H', 'R', 'P', 'F', 'S', 'S', 'S', 'Q', 'K', 'R', 'F', 'I', 'T', 'S', 'L', 'P', 'S', 'R', 'F', 'P', 'T', 'P', 'P', 'L', 'F', '*', 'S', 'P', '*', 'P', 'F', 'C', 'L', 'L', 'E', 'N', 'F', 'I', '*', 'N', 'R', 'I', 'R', 'M', 'G', 'A', 'V', 'A', 'H', 'A', 'C', 'T', 'L', 'A', 'H', 'A', 'C', 'T', 'L', 'G', 'G', 'R', 'G', 'G', 'R', 'I', 'T', '*', 'G', '*', 'E', 'F', 'Q', 'N', 'S', 'V', 'A', 'N', 'V', 'V']
mutations= {'c': 't', 'a': 't', 't': 't', 'g': 't'}
p53_seg_mut= [mutations.get(p53_seg[i]) for i in range(0, len(p53_seg), 100)]
mutation3 = list(p53_seg)
for i in range(0, len(p53_seg_mut)):
mutation3[i*100] = p53_seg_mut[i]
codon3=["".join(mutation3[(3*s):(3*s)+3]) for s in range(0, len((mutation3))/3)]
print codon3
trans3=[standard.get(c) for c in codon3]
print trans3
IDLE: ['W', 'S', 'S', 'S', 'L', 'F', 'T', 'R', '*', 'E', 'G', 'R', 'R', 'E', 'R', 'E', 'N', 'V', 'L', '*', 'A', 'G', 'S', 'S', 'Y', 'L', 'A', 'E', 'G', 'G', 'C', 'Y', 'S', 'L', 'P', 'A', 'F', 'L', 'F', 'L', 'D', 'Y', 'L', 'V', 'M', 'A', 'F', 'A', 'K', 'A', 'G', 'V', 'F', 'V', 'L', 'M', 'Q', 'T', 'S', 'I', 'P', 'P', 'L', 'L', '*', 'M', 'V', 'C', 'P', 'T', 'P', 'R', 'V', 'A', 'C', 'N', 'L', 'G', 'G', 'R', 'Y', 'H', 'G', 'V', 'D', 'R', 'E', 'G', 'K', 'K', 'C', 'A', 'E', 'G', 'K', 'P', 'G', 'G', 'T', 'F', '*', 'N', 'E', 'H', 'I', 'S', 'S', 'S', 'R', 'R', 'K', 'K', 'K', 'K', 'N', 'G', 'T', 'S', 'E', 'N', 'E', 'I', 'L', 'K', 'E', 'C', 'N', 'D', 'G', 'S', 'F', 'D', 'N', 'L', 'S', 'G', 'K', 'T', 'I', 'Y', 'L', 'L', 'S', 'S', 'F', 'G', 'L', 'G', 'H', 'S', 'S', 'S', 'R', 'R', 'R', 'L', 'N', 'V', 'V', 'K', 'R', 'K', 'G', 'R', 'A', 'R', 'S', 'R', 'P', 'C', 'G', 'P', 'P', 'C', 'S', 'P', 'R', 'P', 'E', 'P', 'V', 'L', 'P', 'G', 'R', 'R', 'R', 'N', 'S', 'N', 'S', 'F', 'L', 'P', 'L', 'P', 'H', 'L', 'L', 'A', 'C', 'G', 'C', 'F', 'I', 'P', 'Q', 'F', 'L', 'P', 'M', 'H', 'L', 'P', 'R', 'T', 'G', 'H', 'F', 'V', 'P', 'Y', 'L', 'R', 'H', 'L', 'F', 'P', 'K', 'S', 'R', 'W', 'H', 'L', 'H', 'T', 'A', 'P', 'V', 'H', 'T', 'A', 'S', 'A', 'Q', 'E', 'D', 'E', 'F', 'W', 'P', 'L', 'T', 'A', 'P', '*', 'C', 'L', 'P', 'S', 'H', 'R', 'P', 'F', 'S', 'S', 'S', 'Q', 'K', 'R', 'F', 'I', 'T', 'S', 'L', 'P', 'S', 'R', 'F', 'P', 'T', 'P', 'P', 'L', 'F', '*', 'S', 'P', '*', 'P', 'F', 'C', 'L', 'L', 'E', 'N', 'F', 'I', '*', 'N', '*', 'I', 'R', 'M', 'G', 'A', 'V', 'A', 'H', 'A', 'C', 'T', 'L', 'A', 'H', 'A', 'C', 'T', 'L', 'G', 'G', 'R', 'G', 'G', 'R', 'I', 'T', '*', 'G', '*', 'E', 'F', 'Q', 'I', 'S', 'V', 'A', 'N', 'V', 'V']
SEPTEMBER 15TH: Calculate the Exponential and Logistic curves in Spreadsheet and Python forms as discussed in class (ppt class slides posted if you need it). Please put a brief comment on your wiki page indicating observations that you had in doing this exercise, e.g. an example plot or any problems you encountered.
- Observations: This was a very interesting activity for me, since I'm quite new to programming. I've used two programming languages before (one I don't remember, and ActionScript (circa 2005)), and getting acquainted with Python proved to be a challenge. I first tried to write code for the exponentials using commands other than the one's provided for us by Dr. Church and I got a crazy error message:
File "C:\Python26\Biophysics Ex 1", line 5, in <module>
import matplotlib.pyplot
File "C:\Python26\lib\site-packages\matplotlib\pyplot.py", line 76, in <module>
new_figure_manager, draw_if_interactive, show = pylab_setup()
File "C:\Python26\lib\site-packages\matplotlib\backends\__init__.py", line 25, in pylab_setup
globals(),locals(),[backend_name])
File "C:\Python26\lib\site-packages\matplotlib\backends\backend_wxagg.py", line 23, in <module>
import backend_wx # already uses wxversion.ensureMinimal('2.8')
File "C:\Python26\lib\site-packages\matplotlib\backends\backend_wx.py", line 116, in <module>
raise ImportError(missingwx)
ImportError: Matplotlib backend_wx and backend_wxagg require wxPython >=2.8
When I finally downloaded a version of matplotlib.pyplot that was compatible with Python 2.6.2 and tried out the command provided by Dr. Church, things improved drastically.
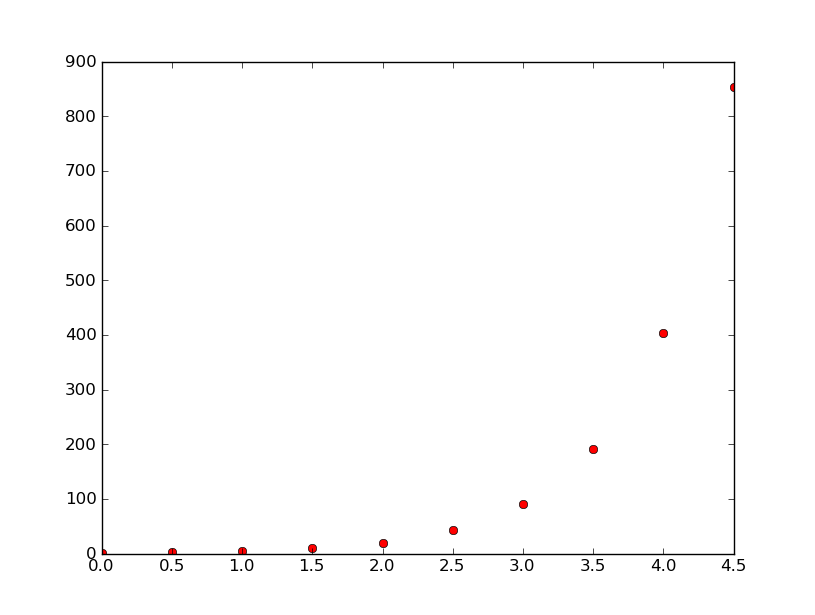
In the spirit of making observations about the exponentials themselves, however: one notes that as the value of k increases from 0.9 to 1.5, considering the same range of x (2.5<x<3.0), that for k=0.9, 10<y<15; for k=1.5, 30<y<100; and for k=3.0, 100<y<1000. These data demonstrate that as the exponential growth constant increases, so does rate of exponentiation. Moreover, using excel to plot the exponentials, I found that for k=3 and using A3=MAX(k*A2*(1-A2), 0) that the plot appeared to take on something of a logistic curve (plus some junk near the top.) Plots for k=3.67859, 4, and 4.03 didn't have good fits for logistic curves.
- Example: k=1.5