User:Perry/Summer 2006 Harvard iGEM/Progress over vacation
BioBricks assembly
The final constructs
- R0010-B0032-Lpp29-OmpA66-Streptavidin
- R0010-B0032-Lpp29-OmpA159-Streptavidin
The parts
R0010 is a Lac promoter, B0032 is a ribosome binding site (medium strength, same one used in the GFP expression device), Lpp29-OmpA66 and Lpp29-OmpA159 are two versions of the membrane display scaffold.
There are several different versions of streptavidin that may be used. We have BioBricks of strepavidin clones from the Ting lab: wild-type, wild-type + His6tag, and dead, which I refer to as StrepW, StrepH, and StrepD.
Available sequenced and midiprepped parts
In Lpp-OmpA box...
- Lpp29, OmpA66, OmpA159 in pCR-2.1 Topo vector
- Lpp29, OmpA66, OmpA159 in pSB1A2 vector
- Lpp29-OmpA66 in pSB1A2
In StrepW,H,D box...
- StrepW, StrepH, StrepD in pCR-2.1 Topo vector
Available digested and gel purified parts, ready for ligation
- R0010, S/P (CIPed)
- Lpp29, E/S
- Lpp29, S/P (CIPed)
- OmpA66, X/P
- OmpA159, X/P
- Lpp29-OmpA66, S/P (CIPed)
- StrepW, StrepH, StrepD, X/P
- pSB1A2, E/P ("1")
- pSB1A2, E/S ("2")
- pSB1A2, X/P ("3")
BioBricks PCR
We're working on getting BioBricks of single-chain dimeric streptavidin clones from the Aslan lab: SCD-NM (single-chain dimer, not mutant), C2, and E2 (two mutant clones), which I refer to as S, C, and E. I just received a core streptavidin (streptavidin has a full-length version and a functional truncated core version) from the Dr. Takeshi Sano, called pTSA13, which I refer to as pTSA13 or just 13.
There are also two clones of the Lpp-OmpA scaffold which I received recently from the Georgiou lab, 481 (Lpp1-9-OmpA46-66-Bla) and 488 (Lpp1-9-OmpA46-145-Bla).
Goals for week of 8/6/06
Assembly
- Lpp29-OmpA159
- Lpp29-OmpA66-StrepW, H, or D
- R0010-B0032
BioBrick PCR
- SCD-NM, C2, or E2 clones of single-chain dimeric streptavidin from Aslan lab, with PstI site removed by mutagenesis
- pTSA13 streptavidin clone from Sano lab
- 481 or 488 construct of Lpp-OmpA from Georgiou lab
Agenda
Monday
- Inoculate liquid cultures, 3ml LB + 30ul amp(5mg/ml), of R0010 and B0032. You can use a R0010 streak on a carb plate labeled 7/31/06 in the B2 refrigerator, and a B0032 streak on a carb plate labeled 8/2/06. Make sure you start incubating/shaking them late enough in the day that you'll be around in 16 hours the following day to perform the miniprep or to take out and refrigerate/pellet for later miniprep.
- Prepare 25ul double digests using midipreps of Lpp29 (SpeI/PstI), OmpA159 (XbaI/PstI), and Lpp29-OmpA66 (SpeI/PstI) in pSB1A2 (Lpp-OmpA box). You should aim for around 1ug of DNA in each digest.
- X ul midiprep for ~1ug DNA, 2.5ul Buffer2, 0.5ul SpeI/XbaI, 0.5ul PstI, 0.25ul BSA(100X), Y ul water for total volume of 25ul
- Y should equal 21.25 minus X. Don't worry about having exactly 0.25ul BSA; just convince yourself there's a little bit of BSA on the end of your pipet tip, and that it's entering your mixture.
- Incubate at 37dC overnight (12h or so), 80dC for 20min, 4dC forever. There should be a program called "Digest" on either of the two grey PCR machines; just change the 37dC step to whatever time.
- X ul midiprep for ~1ug DNA, 2.5ul Buffer2, 0.5ul SpeI/XbaI, 0.5ul PstI, 0.25ul BSA(100X), Y ul water for total volume of 25ul
- Prepare three PCRs using "S/C/E mut in pET" minipreps and StrepSCDF/StrepSCDRS(5uM) primers in "S,C,E" box, and PCR Supermix in "Primers/Stuff" box.
- 45ul PCR Supermix + 2ul StrepSCDF(5uM) + 2ul StrepSCDRS(5uM) + 1ul "S, mut in pET" miniprep
- " + " + " + 1ul "C, mut in pET" miniprep
- " + " + " + 1ul "E, mut in pET" miniprep
- Incubate using "PCR PT" program on either grey PCR machine.
- Get primers Lpp9F and OmpA145R from Alain. Reconstitute in (10 times #nmoles) ul EB for a concentration of 100uM. You should vortex for a good 30-40 seconds. Then dilute 5ul of that with 95ul EB for a dilution of 5uM. Then prepare two PCR reactions with the 481 and 488 minipreps in "Misc." box. Also, the OmpA66R primer should be in the "Primers" box at the bottom of the B5 freezer.
- 45ul PCR Supermix + 2ul Lpp9F(5uM) + 2ul OmpA66R(5uM) + 1ul 481 miniprep
- 45ul PCR Supermix + 2ul Lpp9F(5uM) + 2ul OmpA145R(5uM) + 1ul 488 miniprep
Tuesday
- (Person A - Tiff √) Treat the Lpp29 and Lpp29-OmpA66 digests with alkaline phosphatase (CIP). To each of the 25ul digests, add 1ul alkaline phosphatase and 3ul 10X phosphatase buffer. Incubate at 37dC for one hour, 4dC forever.
- Tiff: Accidentally CIPed the OmpA159 digest too. Perry: Just throw it out and use an X/P OmpA159 I purified 7/27.
- (Person B - Matt √) While you're waiting for CIP, pour a 1% 100ml agarose gel.
- Dissolve 1g agarose in 100ml TBE with microwave at 30-sec intervals. Once the solution is clear, let sit in cold tap water until you can hold the warm bottle comfortably in your hands, then add 5ul EtBr. Swirl until EtBr has disappeared and then pour into a gel frame that has been set even. Use a 15-tooth comb to push all the bubbles down to the bottom of the gel, and then set comb along top. Let gel harden over 15-20min.
- (Person A - Tiff√) While you're waiting for CIP, perform minipreps of R0010 and B0032 liquid cultures from yesterday. Protocol note: I usually include the extra PB step, and elute in 30ul water, letting stand for 5min instead of 1min.
- (Person A - Tiff√) Prepare digests using these minipreps. You can incubate these for 2-3 hours at 37dC and try to include them in the gel for today, or you can incubate them overnight at 37dC and gel isolate/purify them tomorrow. Remember to include the enzyme-killing step of 80dC for 2min at the end, and holding at 4dC.
- 20ul R0010 miniprep + 2.5ul Buffer2 + 1.25ul water + 0.5ul EcoRI + 0.5ul SpeI + 0.25ul BSA(100X)
- 20ul B0032 miniprep + 2.5ul Buffer2 + 1.25ul water + 0.5ul EcoRI + 0.5ul XbaI + 0.25ul BSA(100X)
- You can use 2.5ul BSA(10X) if you want, and then don't add the water and reduce the miniprep amount to 19ul.
- (Person A - Tiff√) Prepare digests using these minipreps. You can incubate these for 2-3 hours at 37dC and try to include them in the gel for today, or you can incubate them overnight at 37dC and gel isolate/purify them tomorrow. Remember to include the enzyme-killing step of 80dC for 2min at the end, and holding at 4dC.
Wednesday
- (Person B Val & Matt √)Perform CIP treatment of B0032 digest. To the 25ul digest, add add 1ul alkaline phosphatase and 3ul 10X phosphatase buffer. Incubate at 37dC for one hour, 4dC forever.
- Pour a 1% 100ml agarose gel while waiting for the CIP if you haven't already.
- (Person B Val & Matt √) Run gel electrophoresis of digests and PCRs.
- Add 3ul loading dye to R0010 and B0032 digests, 4ul loading dye to Lpp29 and Lpp29-OmpA66 digests, 6ul loading dye to PCR reactions "S/C/E, mut in pET" and "481" and "488" from yesterday.
- Load 10ul 1kb+ ladder, entire OmpA159 digest (and R0010 and B0052 if ready) (28ul), entire Lpp29 and Lpp29-OmpA66 digests (33ul), and split up the PCR reactions into two wells for each reaction (28ul in each well).
- Run gel for 45min at 130V. Make sure the gel apparatus is clean and has fresh TBE buffer.
- (Person B Val & Matt √) Image gel and cut out bands. First, prelabel and preweigh empty 1.5ml Eppendorf tubes on the scale in the back with the sliding windows, and record on the tube itself how many milligrams it weighed. Also, set the heatblock to 50dC.
- Expected band sizes, approximate:
- Lpp29, SpeI/PstI: 2200bp
- Lpp29-OmpA66, SpeI/PstI: 150bp (band did not show up; unsuccessful digest?)
- S/C/E PCRs: 850bp and 350bp. Cut out the 850 bp and NOT the 350bp band.
- 481 PCR: 90bp (bands did not show up; unsuccessful PCR)
- 488 PCR: 330bp (bands did not show up; unsuccessful PCR)
- R0010, EcoRI/SpeI: 250bp (band did not show up; unsuccessful digest?)
- B0032, EcoRI/XbaI: 2100bp (band did not show up; unsuccessful digest?)
- Expected band sizes, approximate:
Thursday
- (Matt √) Cut out bands, collect gel slices in tubes, weigh the tubes again and subtract to determine milligrams of gel slice. Use this weight in milligrams as your gel volume for gel extraction.
- (Matt √) Gel extraction. Follow Qiagen protocol, using purple Qiaquick tubes. Protocol note: I include the extra QG step, and elute in 30ul water, letting stand for 5min instead of 1min. There is a small bottle of isopropanol over my bench, B4.
- Handed 7 purified samples (LPP, and S/C/E PCRs) to cyano group Matthewmeisel 16:32, 10 August 2006 (EDT)
- Put these constructs in a newly labeled "PCR products" box in Perry's freezer"--Zsun 21:38, 10 August 2006 (EDT)
- Handed 7 purified samples (LPP, and S/C/E PCRs) to cyano group Matthewmeisel 16:32, 10 August 2006 (EDT)
- Ligation/Topo cloning. This is a rather large undertaking (five ligations, four Topo clonings), so it might be helpful to split it up between two people.
- Samples needed
- Lpp29, SpeI/PstI, CIPed
- OmpA159, XbaI/PstI, purified 7/27
- Lpp29-OmpA66, SpeI/PstI, CIPed Note: didn't work from yesterday, have older miniprepped version: L066 S/P from gel purified box--Zsun 18:38, 10 August 2006 (EDT)
- StrepW, StrepH, and StrepD, XbaI/PstI (in the "gel-purified digests" box)
- S/C/E PCRs from yesterday, gel-purified today
- pTSA13 PCR, gel-purified 8/4/06 (in pTSA13 box, labeled "lanes 2+3")
- R0010, EcoRI/SpeI Note: didn't work from yesterday, don't have this--Zsun 18:38, 10 August 2006 (EDT)
- B0032, EcoRI/XbaI, CIPed Note: didn't work from yesterday, don't have this--Zsun 18:38, 10 August 2006 (EDT)
- (Peng √)Ligations
- 2.5ul Lpp29 S/P + 7.5ul OmpA159 X/P (7/27)
- 2.5ul Lpp29-OmpA66 S/P + 7.5ul StrepW, X/P Note: using old vers--Zsun 18:38, 10 August 2006 (EDT)
- 2.5ul Lpp29-OmpA66 S/P + 7.5ul StrepH, X/P Note: using old vers--Zsun 18:38, 10 August 2006 (EDT)
- 2.5ul Lpp29-OmpA66 S/P + 7.5ul StrepD, X/P Note: using old vers--Zsun 18:38, 10 August 2006 (EDT)
- 2.5ul B0032 E/X + 7.5ul R0010, E/S Note: Won't be able to do--Zsun 18:38, 10 August 2006 (EDT)
- Mix these, add 10ul "Vial 1" ligase buffer (found in "primers/stuff" box, or as part of Rapid DNA ligase kit), and mix again. Then add 1ul "Vial 3" DNA ligase, and mix thoroughly. Let sit at room temperature 10min.
- ( Jeff √) Topo clonings. Use TOPO-TA cloning kit. I would suggest opening a new kit, which should be in the -80 freezer. I was having trouble with my last Topo cloning.
- 4ul gel-purified PCR "S"
- 4ul gel-purified PCR "C"
- 4ul gel-purified PCR "E"
- 4ul gel-purified PCR "pTSA13" Note: labeled PT lanes 2,3 from the pst box--Zsun 18:48, 10 August 2006 (EDT)
- Mix each of these with 1ul salt solution and 1ul Topo vector. Let sit at room temperature 10min.
- Samples needed
- (Peng and Jeff √) Transformations. You can refrigerate the ligation/Topo cloning reactions for later if you don't have time to do the transformation today. Again, it's a large undertaking, so it might be better with two people.
- You'll be doing nine transformations: the five ligations and four Topo clonings from above, and of course you're not going to use nine tubes of Top10 cells (they're about $15 a pop). I would thaw on ice (takes 5min or so) three tubes of Top10 cells (in the -80 freezer, bottom shelf, labeled "Top10 chemically competent cells"), and then aliquot 20ul into 9 tubes.
- You can include a positive control (transform with 2ul of pUC19 DNA, an amp-resistance plasmid, found in the same box as the Top10 cells) or negative control (transform with water or nothing). I've been consistently getting good positive/negative controls with the transformation protocol, and so have stopped including the controls in order to save cells and plates; but if you feel less confident, please include them. That would make 9+2=11 transformations, and you should aliquot ~15ul instead of 20ul. The rest of the protocol should be exactly the same for these transformations, using carb/amp plates.
- After you've aliquotted the cells into different tubes, pipet in the respective 21ul ligation reaction or 6ul Topo cloning (or 2ul pUC19), tap or flick the tubes a bunch (that's the SI unit for 10ish) of times to mix, and leave the cells on ice for 30min or longer if you're on lunch break or something (I'd say no more than an hour). While you're waiting, make sure the heatblock is at 42dC, and squirt some water in the holes for better heat contact during the heat shock step, which is next. Also, if you're around during this incubation-on-ice step, it doesn't hurt to tap or flick the tubes a few times every ten or fifteen minutes to help the transformation by mixing.
- Heat-shock the cells at 42dC on heatblock for 30 sec, then cool on ice for at least 2min. Add 200ul SOC media to each tube, and leave in the 37dC shaker for 1 hour or more (I'd say no more than an hour and a half). While they're shaking, leave five carb plates (for ligations; add two for controls) and four kan plates (for Topo clonings) to warm in the plates incubator.
- If there are no kan plates, you can use carb/amp plates as well for Topo clonings because the Topo vector is both amp and kan resistant.
- Then under a flame, pipet the whole culture onto an appropriately labeled plate and spread with a blue plastic spreader, and leave the plates agar-side-up in the plates incubator for 15-16 hours.
- Make sure you plate the cultures late enough in the day that someone will be here 15-16 hours later to take them out of incubation. You may refrigerate the SOC media mixtures after their one-hour incubation until they're ready for plating.
- There are going to be 10 plates; 1 neg, 1 pos, 4 ligation, and 4 topo. four are kan and six are carb, where the kan are for Topo. Sitting in the 37 crirca 930PM.--Zsun 21:38, 10 August 2006 (EDT)
- Make sure you plate the cultures late enough in the day that someone will be here 15-16 hours later to take them out of incubation. You may refrigerate the SOC media mixtures after their one-hour incubation until they're ready for plating.
- You'll be doing nine transformations: the five ligations and four Topo clonings from above, and of course you're not going to use nine tubes of Top10 cells (they're about $15 a pop). I would thaw on ice (takes 5min or so) three tubes of Top10 cells (in the -80 freezer, bottom shelf, labeled "Top10 chemically competent cells"), and then aliquot 20ul into 9 tubes.
Friday
- (Person A - Peng √)'
- You have eight transformation plates, each with colonies on them we hope. A positive control plate should have plenty of colonies; a negative control plate should have no colonies. Remove all plates from the incubator.
- The colonies should be sizeable white dots, about .5-1 millimeter in diameter, big enough to pick from. If the colonies aren't large enough, leave the plates in for another hour or so. If they're overgrown, that is, if there are "satellite colonies" which are smaller white dots surrounding the large colonies, make sure that when you pick from the centers of the large colonies. (The satellite colonies are often bacteria without the correct resistance which are growing due to the depletion of drug concentration by the growth of the larger colonies.)
- Ligation assemblies in pSB1A2 vector
- Lpp29-OmpA159
- Lpp29-OmpA66-StrepW
- Lpp29-OmpA66-StrepH
- Lpp29-OmpA66-StrepD
- Inserts in Topo vector
- "S" strep clone with PstI site removed
- "C" strep clone with PstI site removed
- "E" strep clone with PstI site removed
- "pTSA13" strep clone
- Preparing for streak/colony PCR (Person A - Peng √)'
- Warm up one carb plate by leaving it in the 37dC plates incubator.
- Prepare two colony PCR mixes.
- The VF2 and VR primers flank the insert site in the pSB1A2 vector. You'll be doing colony PCR to check four ligations, so it's a good idea to make enough master mix for five PCRs, so you don't run out. A 10ul PCR reaction is good enough, and you want about 200nM of each primer, so in one tube, mix 40ul PCR Supermix + 5ul VF2(2uM) + 5ul VR(2uM). Then aliquot 10ul into four tubes, each labeled for its respective ligation assembly check.
- The M13F(-21) and M13R primers flank the insert site in the Topo vector. You'll be doing colony PCRs to check four inserts, so make enough master mix for five PCRs. In one tube, 40ul PCR Supermix + 5ul M13F(-21)(2uM) + 5ul M13R(2uM). Then aliquot 10ul into four tubes, each labeled for its respective Topo insert check.
- PCR Supermix and primers can be found in my "Primers/Stuff" box. If you run out of Supermix, there should be more in the 5096 -20dC freezer.
- Remove the carb plate from the incubator, wipe off the outside condensation, and with a marker on the outside of the agar side, draw lines to divide the plate up into eight sections, and label the sections according to the ligation assemblies and Topo inserts.
- Streak/colony PCR. (Person A - Peng √)'
- Do this under a flame. Run a plastic yellow inoculation needle (there are some in the drawer at my bench B4; there's a box labeled "Nunc" in 5096) through the flame, then pick a single colony from the transformation plate, and streak along the agar in the labeled section on the empty carb plate (note that you'll be reading the section labels backwards), and then dip the same needle into the PCR mix in the correspondingly labeled tube and twist/wiggle it around to make sure some of the bacteria entered the PCR mix. Leave the streak plate in the 37dC plates incubator, at an appropriate time that someone will be around in ~16hours to take the plate out. Otherwise, leave the plate in 4dC refrigeration. Place tubes in one of the grey PCR machines and run "PCR PT" program.
- Run PCRs on 1.2% e-gel. (Person A - Peng √)'
- Run 1kb+ ladder in one of the lanes (10ul 1kb+ ladder plus 10ul water). For the PCRs, add 10ul water to the PCR mixtures, and then load the 20ul into the wells. Otherwise, follow the e-gel protocol for operation. Remember to load empty wells with 20ul water.
- Expected band sizes for PCRs. Note: The VF2/VR and M13F/M13R primer pairs both anneal approximately 100bp upstream and downstream of the insert site, so band sizes will be ~200bp larger than the actual insert.
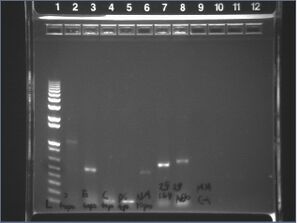
- Lpp29-OmpA159 (VF2/VR): 630bp
- Lpp29-OmpA66-StrepW (VF2/VR): 640bp
- Lpp29-OmpA66-StrepH (VF2/VR): 660bp
- Lpp29-OmpA66-StrepD (VF2/VR): 640bp
- "S" strep clone with PstI site removed: 1080bp
- "C" strep clone with PstI site removed: 1080bp
- "E" strep clone with PstI site removed: 1080bp
- "pTSA13" strep clone: 610bp
- Inoculate cultures from clones that yielded the correct band sizes.
- NOTE: solid plate in the incubator circa 230PM Friday; will take out sat. and do the innoculation step.
- If you got the correct band size from a colony PCR, you should inoculate a culture that will be miniprepped, and the miniprepped DNA will be sequenced. Label a plastic culture tube (some in a drawer at my bench B4) according to its ligation/Topo cloning, and under a flame, pipet 3ml LB and 30ul amp(5mg/ml) or 3ul amp(50mg/ml). If you have a bunch of inoculations to do, make a master mix (enough for n+1 cultures) in a 50ml tube of LB+amp.
- Do this under a flame. Run a plastic yellow inoculation needle through the flame, and then pick from the end of the streak you made earlier (it might be hard to see, but the bacteria should be there). Dip the same needle into the LB+amp mixture in the tube, and shake it around a little bit to make sure the bacteria entered the LB mixture.
- Leave the liquid culture tubes in the 37dC incubator/shaker, late enough that someone will be around in 12-16hours to take them out and pellet/freeze them. Otherwise, wait on the inoculations for later.
- Later, these liquid cultures would be miniprepped and then sent for sequencing.
- If you have time, you can try to streak and colony PCR from transformations that didn't yield the correct band size. Just repeat the process with other colonies on the transformation plate.
Update
The plate divided into 6 grew a bit too well... restreaked that onto another plate, and put in the 37 around 5PM Sat; also restreaked a plate marked MM 8/7 PT and grew at same time. This can then be refrigerated and the project continued by the next person.--Zsun 17:39, 12 August 2006 (EDT)
Over the weekend/next week
- If you have time, repeat the streak/colony PCR/e-gel process with other colonies on the C, E, and pTSA13 transformations plates, maybe picking from two or three colonies this time. You may streak on an amp or a kan plate since all of these are in amp/kan-resistance Topo vector.
- It might prove more efficient to do this before you move on to the following steps, so as to be able do a couple more digest checks or prepare a couple more sequencing reactions in parallel. Please read all of the following once through first, b/c several things can be done at the same time.
- The band size is correct for S in Topo; however, a digest check needs to be done first to make sure that the PstI site has been removed. Inoculate 3ml LB cultures and miniprep, but elute with 40ul water. Use 20ul of that miniprep in the following digest. (If you found successful C and E Topo inserts by colony PCR, they would be included here.)
- 20ul miniprep + 2.5ul Buffer2 + 1.25ul water + 0.5ul XbaI + 0.5ul PstI + 0.25ul BSA(100X).
- 2.5ul BSA(10X) may be used. If you do, do not add water, and use 19ul miniprep, to maintain a total volume of 25ul.
- Incubate at 37dC for a few hours or overnight, depending on your schedule, then 80dC for 20min. Then run on an agarose gel or e-gel. If using an e-gel, load only 10ul digest + 10ul water, so as not to overload the e-gel.
- If there is a band ~850bp, then the PstI site has been successfully removed by mutagenesis. Send the remaining miniprep (there should be at least 16ul left) for sequencing.
- If you ran an agarose gel, you might as well excise and gel-extract this 850bp fragment, since it may be used later in suffix insertion assemblies.
- 20ul miniprep + 2.5ul Buffer2 + 1.25ul water + 0.5ul XbaI + 0.5ul PstI + 0.25ul BSA(100X).
- It looks like the band sizes are correct for Lpp29-OmpA159 and Lpp29-OmpA66-StrepD (Peng, I realized I had forgotten to add 200bp for the Lpp29-OmpA66-Strep band sizes). Inoculate liquid cultures from those streaks, miniprep, then send out for sequencing. Note that you can't make a sequencing order until Monday or later, so plan accordingly. (If you found a successful pTSA13 insert by colony PCR, that would be included here.)
- There's one other clone that I wanted to sequence as well, from a while ago. This is H#5, which should be on a many-streak plate from 8/3/06. It's StrepH in pSB1A2, and the #5 means that it was the 5th colony that I tried to check for insert by colony PCR. Inoculate a 3ml LB culture, miniprep, and send out for sequencing.
- For sequencing, a forward sequencing reaction and a reverse sequencing reaction should be ordered for each construct, and 8ul miniprepped DNA should be provided for each reaction. Constructs in Topo vector can have M13F(-21) and M13R primers added by Genewiz; you have to add 4ul of VF2 and of VR primers (2uM) for constructs in pSB1A2 vector.
- It would be good to revisit the assemblies that got derailed along the way: R0010-B0032, Lpp29-OmpA66-StrepW, and Lpp29-OmpA66-StrepH.
- You can try repeating the LO66W and LO66H ligation/transformations with different volume ratios that better reflect a 1:3 vector:insert molar ratio. Use the nanodrop to calculate the mass concentrations of each gel extraction and dimensional analysis to convert to molar concentration. The molar weight of the Lpp29-OmpA66 in pSB1A2 vector is ~1,400,000 g/mol, and the molar weights of the StrepW and StrepH fragments are each ~260,000 g/mol. Then try approximating volumes that yield a 1:3 molar ratio; they can add to 10ul, or add to less than 10ul, in which case you'd need to add DNA dilution buffer from "Vial 2" in the Rapid DNA ligase kit to fill up to 10ul.
- For the R0010-B0032 assembly, you would need to reinoculate R0010 and B0032 cultures from their respective streaks, then prepare digests as previously described.