User:Mbennie/Notebook/Lab Notebook/Notebook/2007/08/04
From OpenWetWare
- PCR Purification
- Used MinElute columns to purify B, C, D, and IgAbc complete (no muts) PCR product from yesterday
- Eluted in 10ul of water
- Colony PCR
- Template: 20ul Supermix, .8ul VF2 and VR, 1 ul of cell dilution (colony in 100ul of water)
- Picked three colonies from each plated sample to PCR (Signal Sequence, IgAbc R, Fos, JunB, GCN4)
- Plated picked colonies on Amp/Cl plate and grew up at 37C all day and overnight
- Protocol:
- 95C for 15 mins
- 94C for 30 secs
- 55C for 30 secs
- 68C for 1 min
- REPEAT 2-4 39 times
- 68C for 10 mins
- 4C FOREVER
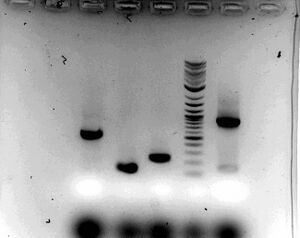
- Gel
- Ran 1% for 30 minutes at 100V to ensure that PCR worked
- Everything looks good
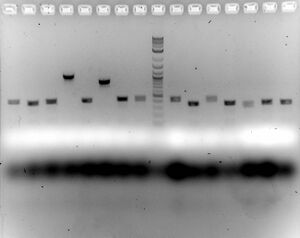
- Gel
- Ran 1.5% for 50 minutes at 100V to see if PCR product is correct length
- Expected sizes (without VF and VR2 additions): Signal Sequence (140 bp), IgAbc (892 bp), Fos (183 bp), JunB (180 bp), GCN4 (156 bp)
- Looks like we might have some correct inserts
- Onwards to sequencing
- Liquid Cultures
- Grew up samples in 8ml of Amp/Cl media for sequencing overnight in 37C incubator (inoculated with colony PCR cell dilution):
- Signal Sequence #2
- Signal Sequence #3
- IgAbc #1
- Fos #1
- Fos #3
- JunB #3
- GCN4 #2
- Grew up samples in 8ml of Amp/Cl media for sequencing overnight in 37C incubator (inoculated with colony PCR cell dilution):