User:Justin Roth Muehlmeyer/Notebook/307L Notebook/Poisson
Set Up
Searching for the cosmic radiation signals:
Using a Tektronix 485 Oscilloscope (analog) and ORTEC model # 266 PM base.
Dynode output on PMT to oscilloscope Input Voltage -1500 V PMT outside of lead brick cave
Analog Oscilloscope settings:
- 5 mV/division
- 1 ms/division
- AC triggering: negative slope (internal)
We observe occasional peaks per second that are distinguishable from noise. They change so fast that we cant actually measure the amplitude of their peaks. This is our presumed cosmic radiation.
Using an amplifier:
- We addded a ORTEC amplifier to our board, and connected the output of the diode to the input of the amplifyer. We wanted to test the signal coming out of the amplifier with the oscilloscope before connecting it into the computer. We ran into many difficulties even finding our cosmic radiation signal with the amplifier, and then again while bypassing it.
We had extensive difficulties in trying to get a signal of the cosmic radiation using the amplifier.
Troubleshooting:
New amplifier: Canberra PAD 814.
Switched from dynode to the anode on the PMT, with no effect.
Switched to new PMT - Tracor Northern Model # TN1222. Way clearer, greater magnitude cosmic radiation spikes with just the oscilloscope. Input ~-1400 V. Positive V output.
Still no results from the amplifier, so...
New Oscilloscope: Tektronix TDS 1002 (digital).
"Derivative signal" from the amplifier. Set up: PMT to PREAMP IN, PREAMP switch to IN, AMP OUT to oscilloscope (DC coupling, rising slope, 36.0 mV)
Hooking it up to the software (MCA):
Software: PCA3 in MSDOS Bins: 512 We decided to first take our dwell time to be 1 second.
Other passes: dwell time of 1, 100, 10000 microseconds,
Data
The data we received in the computer was outputed to ASCII file type, which I will post here. The following was the data taken for the 1s trial.
Sep 22 2008 4:01:29 pm Elt: 000000 Seconds. Real Time: 000000 ID: No spectrum identifier defined Memory Size: 16384 Chls Conversion Gain: 0256 Adc Offset: 0000 Chls
Chn Counts ROI 0, 16, 000 1, 16, 000 2, 12, 000 3, 12, 000 4, 13, 000 5, 12, 000 6, 14, 000 7, 11, 000 8, 12, 000 9, 11, 000 10, 13, 000 11, 14, 000 12, 15, 000 13, 14, 000 14, 13, 000 15, 14, 000 16, 17, 000 17, 14, 000 18, 14, 000 19, 14, 000 20, 14, 000 21, 14, 000 22, 14, 000 23, 11, 000 24, 16, 000 25, 13, 000 26, 10, 000 27, 15, 000 28, 17, 000 29, 11, 000 30, 13, 000 31, 11, 000 32, 12, 000 33, 14, 000 34, 14, 000 35, 13, 000 36, 11, 000 37, 12, 000 38, 13, 000 39, 13, 000 40, 16, 000 41, 13, 000 42, 14, 000 43, 11, 000 44, 15, 000 45, 16, 000 46, 13, 000 47, 11, 000 48, 12, 000 49, 11, 000 50, 16, 000 51, 13, 000 52, 15, 000 53, 15, 000 54, 10, 000 55, 17, 000 56, 14, 000 57, 17, 000 58, 16, 000 59, 13, 000 60, 14, 000 61, 16, 000 62, 13, 000 63, 11, 000 64, 13, 000 65, 16, 000 66, 10, 000 67, 16, 000 68, 12, 000 69, 17, 000 70, 13, 000 71, 11, 000 72, 13, 000 73, 16, 000 74, 13, 000 75, 14, 000 76, 12, 000 77, 14, 000 78, 14, 000 79, 13, 000 80, 12, 000 81, 11, 000 82, 15, 000 83, 15, 000 84, 13, 000 85, 14, 000 86, 10, 000 87, 14, 000 88, 15, 000 89, 20, 000 90, 13, 000 91, 14, 000 92, 10, 000 93, 14, 000 94, 13, 000 95, 12, 000 96, 12, 000 97, 14, 000 98, 15, 000 99, 14, 000 100, 14, 000 101, 13, 000 102, 13, 000 103, 11, 000 104, 16, 000 105, 14, 000 106, 9, 000 107, 18, 000 108, 9, 000 109, 15, 000 110, 11, 000 111, 16, 000 112, 14, 000 113, 12, 000 114, 15, 000 115, 16, 000 116, 16, 000 117, 13, 000 118, 14, 000 119, 16, 000 120, 16, 000 121, 13, 000 122, 15, 000 123, 15, 000 124, 12, 000 125, 16, 000 126, 11, 000 127, 13, 000 128, 14, 000 129, 15, 000 130, 15, 000 131, 10, 000 132, 12, 000 133, 13, 000 134, 15, 000 135, 12, 000 136, 16, 000 137, 14, 000 138, 12, 000 139, 15, 000 140, 19, 000 141, 13, 000 142, 15, 000 143, 11, 000 144, 16, 000 145, 16, 000 146, 13, 000 147, 13, 000 148, 13, 000 149, 15, 000 150, 14, 000 151, 13, 000 152, 14, 000 153, 12, 000 154, 14, 000 155, 15, 000 156, 15, 000 157, 14, 000 158, 13, 000 159, 16, 000 160, 12, 000 161, 15, 000 162, 14, 000 163, 11, 000 164, 15, 000 165, 8, 000 166, 10, 000 167, 15, 000 168, 17, 000 169, 11, 000 170, 15, 000 171, 11, 000 172, 12, 000 173, 15, 000 174, 18, 000 175, 12, 000 176, 11, 000 177, 11, 000 178, 10, 000 179, 15, 000 180, 14, 000 181, 12, 000 182, 17, 000 183, 10, 000 184, 18, 000 185, 13, 000 186, 15, 000 187, 14, 000 188, 14, 000 189, 15, 000 190, 12, 000 191, 17, 000 192, 10, 000 193, 11, 000 194, 14, 000 195, 14, 000 196, 11, 000 197, 15, 000 198, 14, 000 199, 13, 000 200, 13, 000 201, 18, 000 202, 10, 000 203, 14, 000 204, 12, 000 205, 11, 000 206, 16, 000 207, 11, 000 208, 11, 000 209, 13, 000 210, 16, 000 211, 12, 000 212, 15, 000 213, 16, 000 214, 12, 000 215, 14, 000 216, 17, 000 217, 14, 000 218, 12, 000 219, 16, 000 220, 15, 000 221, 17, 000 222, 14, 000 223, 16, 000 224, 18, 000 225, 15, 000 226, 15, 000 227, 12, 000 228, 11, 000 229, 13, 000 230, 14, 000 231, 15, 000 232, 14, 000 233, 15, 000 234, 13, 000 235, 17, 000 236, 14, 000 237, 12, 000 238, 13, 000 239, 12, 000 240, 8, 000 241, 13, 000 242, 14, 000 243, 9, 000 244, 14, 000 245, 18, 000 246, 14, 000 247, 12, 000 248, 16, 000 249, 14, 000 250, 15, 000 251, 13, 000 252, 14, 000 253, 14, 000 254, 13, 000 255, 16, 000 256, 14, 000 257, 17, 000 258, 16, 000 259, 13, 000 260, 14, 000 261, 9, 000 262, 15, 000 263, 14, 000 264, 16, 000 265, 13, 000 266, 15, 000 267, 14, 000 268, 12, 000 269, 13, 000 270, 14, 000 271, 13, 000 272, 15, 000 273, 12, 000 274, 14, 000 275, 13, 000 276, 13, 000 277, 13, 000 278, 10, 000 279, 15, 000 280, 16, 000 281, 13, 000 282, 16, 000 283, 13, 000 284, 14, 000 285, 10, 000 286, 13, 000 287, 15, 000 288, 14, 000 289, 13, 000 290, 17, 000 291, 15, 000 292, 11, 000 293, 11, 000 294, 16, 000 295, 14, 000 296, 14, 000 297, 16, 000 298, 9, 000 299, 12, 000 300, 12, 000 301, 15, 000 302, 13, 000 303, 10, 000 304, 14, 000 305, 15, 000 306, 12, 000 307, 11, 000 308, 12, 000 309, 13, 000 310, 16, 000 311, 12, 000 312, 8, 000 313, 15, 000 314, 14, 000 315, 16, 000 316, 10, 000 317, 13, 000 318, 13, 000 319, 12, 000 320, 13, 000 321, 12, 000 322, 17, 000 323, 14, 000 324, 14, 000 325, 14, 000 326, 12, 000 327, 14, 000 328, 16, 000 329, 14, 000 330, 10, 000 331, 14, 000 332, 14, 000 333, 14, 000 334, 14, 000 335, 12, 000 336, 16, 000 337, 13, 000 338, 10, 000 339, 12, 000 340, 12, 000 341, 11, 000 342, 15, 000 343, 16, 000 344, 10, 000 345, 13, 000 346, 13, 000 347, 13, 000 348, 10, 000 349, 14, 000 350, 15, 000 351, 14, 000 352, 13, 000 353, 14, 000 354, 16, 000 355, 14, 000 356, 18, 000 357, 9, 000 358, 13, 000 359, 14, 000 360, 10, 000 361, 11, 000 362, 13, 000 363, 15, 000 364, 15, 000 365, 14, 000 366, 11, 000 367, 17, 000 368, 13, 000 369, 11, 000 370, 15, 000 371, 16, 000 372, 16, 000 373, 13, 000 374, 13, 000 375, 13, 000 376, 11, 000 377, 15, 000 378, 10, 000 379, 11, 000 380, 16, 000 381, 15, 000 382, 16, 000 383, 14, 000 384, 18, 000 385, 13, 000 386, 14, 000 387, 16, 000 388, 14, 000 389, 14, 000 390, 12, 000 391, 12, 000 392, 13, 000 393, 8, 000 394, 18, 000 395, 14, 000 396, 7, 000 397, 13, 000 398, 11, 000 399, 13, 000 400, 11, 000 401, 15, 000 402, 12, 000 403, 14, 000 404, 10, 000 405, 12, 000 406, 16, 000 407, 15, 000 408, 13, 000 409, 14, 000 410, 12, 000 411, 11, 000 412, 13, 000 413, 13, 000 414, 17, 000 415, 13, 000 416, 13, 000 417, 15, 000 418, 13, 000 419, 16, 000 420, 14, 000 421, 12, 000 422, 9, 000 423, 16, 000 424, 12, 000 425, 13, 000 426, 15, 000 427, 14, 000 428, 12, 000 429, 11, 000 430, 16, 000 431, 11, 000 432, 14, 000 433, 16, 000 434, 7, 000 435, 15, 000 436, 15, 000 437, 13, 000 438, 14, 000 439, 12, 000 440, 11, 000 441, 12, 000 442, 13, 000 443, 15, 000 444, 12, 000 445, 15, 000 446, 13, 000 447, 14, 000 448, 13, 000 449, 15, 000 450, 12, 000 451, 11, 000 452, 16, 000 453, 16, 000 454, 15, 000 455, 14, 000 456, 12, 000 457, 12, 000 458, 13, 000 459, 13, 000 460, 12, 000 461, 14, 000 462, 15, 000 463, 13, 000 464, 13, 000 465, 16, 000 466, 14, 000 467, 13, 000 468, 14, 000 469, 15, 000 470, 17, 000 471, 14, 000 472, 13, 000 473, 8, 000 474, 12, 000 475, 15, 000 476, 16, 000 477, 11, 000 478, 13, 000 479, 12, 000 480, 14, 000 481, 13, 000 482, 13, 000 483, 15, 000 484, 14, 000 485, 14, 000 486, 11, 000 487, 14, 000 488, 12, 000 489, 13, 000 490, 14, 000 491, 12, 000 492, 15, 000 493, 11, 000 494, 8, 000 495, 18, 000 496, 14, 000 497, 14, 000 498, 12, 000 499, 11, 000 500, 12, 000 501, 15, 000 502, 10, 000 503, 12, 000 504, 14, 000 505, 16, 000 506, 14, 000 507, 16, 000 508, 14, 000 509, 12, 000 510, 13, 000 511, 15, 000
Results
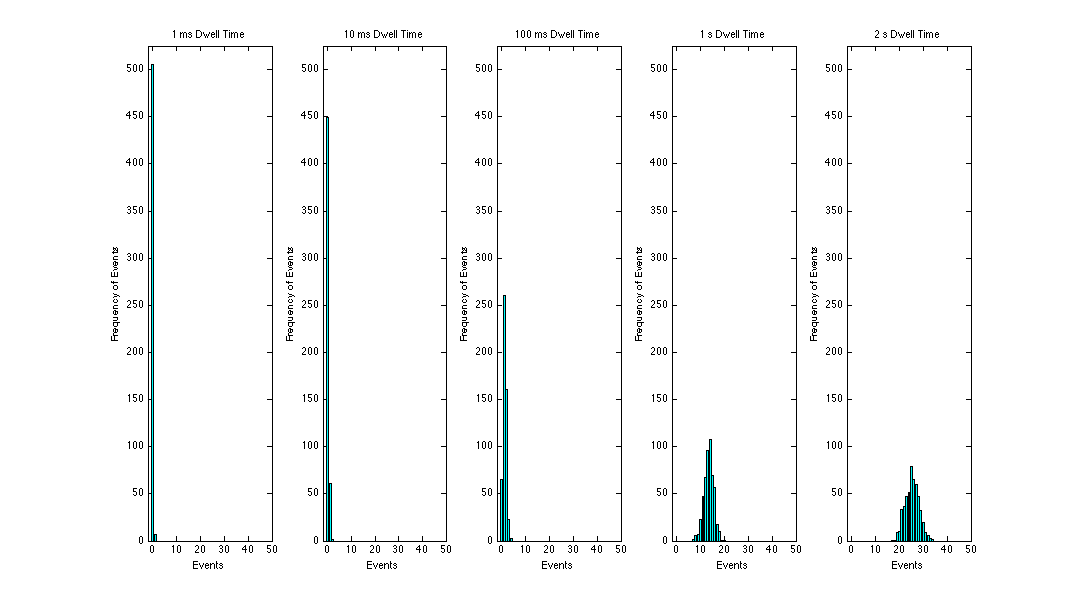
Using Mr. Barron's amazing Matlab skills our results were as follows for each dwell time:
Here is the code for this:
%% Poisson Distribution Analysis
% Alexander Barron
% Junior Lab Fall 2008
close all, clear all;
%% Load .txt files
M1ms_bin = dlmread('JAND1MS.txt',',',[0 0 511 0]); % bin vector 1ms dwell
M1ms_val = dlmread('JAND1MS.txt',',',[0 1 511 1]); % value vector 1ms dwell
M10ms_bin = dlmread('JAND10MS.txt',',',[0 0 511 0]);
M10ms_val = dlmread('JAND10MS.txt',',',[0 1 511 1]);
M100ms_bin = dlmread('JAND100M.txt',',',[0 0 511 0]);
M100ms_val = dlmread('JAND100M.txt',',',[0 1 511 1]);
M1s_bin = dlmread('JAND1S.txt',',',[0 0 511 0]);
M1s_val = dlmread('JAND1S.txt',',',[0 1 511 1]);
M2s_bin = dlmread('JAND2S.txt',',',[0 0 511 0]);
M2s_val = dlmread('JAND2S.txt',',',[0 1 511 1]);
%% sort into frequencies for histograms
d1ms_count = zeros(1,50); % Here I am creating empty
d10ms_count = zeros(1,50); % vectors in which to put
d100ms_count = zeros(1,50); % my manipulated data.
d1s_count = zeros(1,50);
d2s_count = zeros(1,50);
for c=1:5;
switch c;
case 1;
valmat = M1ms_val; % generic value and
countmat = d1ms_count; % count matrices
case 2;
valmat = M10ms_val;
countmat = d10ms_count;
case 3;
valmat = M100ms_val;
countmat = d100ms_count;
case 4;
valmat = M1s_val;
countmat = d1s_count;
case 5;
valmat = M2s_val;
countmat = d2s_count;
end;
for j=1:512; % Here I scan my data for
% desired frequencies
for k=0:50; % and place them in
% generic matrices.
if valmat(j) == k;
countmat(k+1) = countmat(k+1) + 1;
end
end
end
switch c;
case 1;
d1ms_count = countmat; % transformation of
case 2; % generic matrices
d10ms_count = countmat; % into appropriately-
case 3; % named ones
d100ms_count = countmat;
case 4;
d1s_count = countmat;
case 5;
d2s_count = countmat;
end;
clear valmat, clear countmat;
end;
%% Plot
freq = linspace(0,49,50);
scrsz = get(0,'ScreenSize');
figure('Position',[1 scrsz(4)/1.5 scrsz(3)/1.25 scrsz(4)/1.4]);
for f=1:5;
switch f;
case 1;
data = d1ms_count;
titl = '1 ms Dwell Time';
case 2;
data = d10ms_count;
titl = '10 ms Dwell Time';
case 3;
data = d100ms_count;
titl = '100 ms Dwell Time';
case 4;
data = d1s_count;
titl = '1 s Dwell Time';
case 5;
data = d2s_count;
titl = '2 s Dwell Time';
end;
subplot(1,5,f), bar(freq,data,'c');
ylim([0 525]);
xlim([-2 50]);
set(gca,'XTick',[0 10 20 30 40 50]);
ylabel('Frequency of Events');
xlabel('Events');
title(titl);
clear data;
end;
Analysis
The Poisson distribution:
- [math]\displaystyle{ f(k; \lambda)=\frac{\lambda^k e^{-\lambda}}{k!},\,\! }[/math]
Calculating the mean:
In the Poisson distribution, the mean and the variance are the same thing, which is λ. λ is the expected rate of events, the expected number of occurences per time interval. To calculate λ I will use the data average method. I sum up the number of events that occurred, and divide by the number of times I took those events.
- [math]\displaystyle{ \widehat{\lambda}_\mathrm{MLE}=\frac{1}{n}\sum_{i=1}^n k_i. \! }[/math]
My excel file data sheet that I used to do this analysis I will place here:
File:Poisson Analysis for lamda Muehlmeyer.xls
The following is a summary of my λ values for each dwell time using the data average method.
- 1ms, λ = 0.013671875
- 10ms, λ = 0.126953125
- 100ms, λ = 1.294921875
- 1s, λ = 13.48828125
- 2s, λ = 25.36132813
This makes sense, as our dwell time increases, our average rate of events increases. And when you compare each of these values to our plots, they are quite apparently the mean.
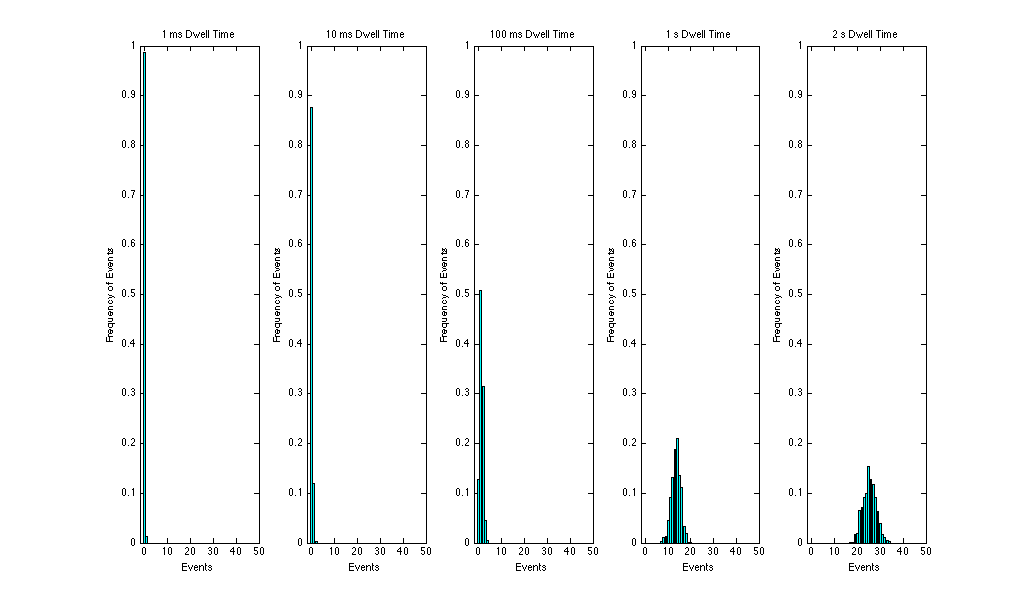
In order to see our results more clearly, we can normalize our results so that they fit a probablity density function that sums to 1. This plots our results as a matter of probabilty. We do this by dividing the frequency of events by the maximimum possibility 512. See the plots below.
What is most interesting is that as we increase the dwell time the histogram plot begins to look like a Gaussian function. The Guassian function peaks at some median value that represents the most common rate of occurences. If we allow for more events (higher dwell time) the histogram then is a plot of this Guassian funtion, which tells us their is a rate of events that occurs the most, and that rate is that at which the Guassian peak appears, our mean, λ.
Error Analysis for λ
SJK 10:34, 20 October 2008 (EDT)

See my comments about this in Der's book
Our error is determined by the deviation from the mean. We found an expression for this value in a formal report from a student from last year. You can see it here at Tomas Mondragon's notebook entry.
[math]\displaystyle{ Error=\frac{\sqrt{\sum_{k=\operatorname{min}\ k}^{\operatorname{max}\ k}\left(f(k,\lambda)-\frac{x_k}{N} \right)^2}}{\sqrt{N}\sqrt{N-1}} }[/math]
Where the frequency count of an event count [math]\displaystyle{ k }[/math] is [math]\displaystyle{ x_k }[/math]. N is 512 in this case.
- 1ms, λ = 0.013671875, λerror = 4.05974132 x 10^-7
- 10ms, λ = 0.126953125, λerror= 1.7313458323 x 10^-5
- 100ms, λ = 1.294921875, λerror= 4.62059546375 x 10^-4
- 1s, λ = 13.48828125, λerror= 3.25904896073 x 10^-4
- 2s, λ = 25.36132813, λerror= 2.4259613547 x 10^-4
Fitting our Data to the Poisson Distribution
The Poisson distribution is characterized by the fact that the standard deviation is equal to the square root of the mean λ. If we can show that this is the case for our data (or atleast reasonably close), then we have shown that our data is in fact a manifestation of the Poisson distribution.
First I will find the standard deviation by assuming that our data is in fact a Poisson distribution (I will take the square root of our calculated λ values):
The number of observed occurences fluctuates about its mean λ with a standard deviation of σk = [math]\displaystyle{ \sqrt{\lambda} }[/math].
- 1ms, λ = 0.013671875, σk = 0.11692
- 10ms, λ = 0.126953125, σk = 0.35650
- 100ms, λ = 1.294921875, σk = 1.13795
- 1s, λ = 13.48828125, σk = 3.67264
- 2s, λ = 25.36132813, σk = 5.03600
This can be roughly validated if one looks at the plots to notice how the distribution deviates out from the mean λ by ±σk.
Now I will determine the standard deviation from our data directly to show that these values for the standard deviation and the values σk above are reasonably close.
This calculation is described by the following formula:
- [math]\displaystyle{ \sigma = \sqrt{\frac{1}{N-1} \sum_{i=1}^N (x_i - \overline{x})^2}\,, }[/math]
Using the excel function for standard deviation (posted above) the standard deviation of our actual data was determined as follows.SJK 10:35, 20 October 2008 (EDT)

This does look correct. However, I don't see the calculations in your actual Excel spreadsheet...just the mean in there.
- 1ms, λ = 0.013671875, σx = 0.116238308
- 10ms, λ = 0.126953125, σx = 0.344790832
- 100ms, λ = 1.294921875, σx = 0.766810045
- 1s, λ = 13.48828125, σx = 2.066378743
- 2s, λ = 25.36132813, σx = 2.945079248
How close are σk and σx?
For low dwell times σx is very close to σk, but as we increase our dwell time the two values begin to differ a bit more. However, I think it is still safe to say that the two values are reasonably close enough to be considered a Poisson distribution.
Why do the two values of σ differ for higher dwell times? Since σ is a measure of deviation of the values from their mean (dispersion), I think that this could be explained by the simple fact that outlyers occur more frequently with longer dwell times, which throw off our data from the ideal Poisson distribution.
Conclusion
The data collection of "random" cosmic radiation events proved to fit the Poisson distribution reasonably well. We also saw how, as we increase the dwell time, this distribution begins to fit a Gaussian with an obvious average rate. We showed our selves that our data is characterized by the fact that the standard deviation is reasonably equal to the square root of the mean λ, which is a charactersistic of the Poisson distribution.