User:Claire E. Lunde/Notebook/Biology 210 at AU
Observation of Zebrafish Embryo Development with Exposure to Rhodamine 03.16.15
Intro: Embryology is the sequence of events, after egg fertilization, involving growth, the structural and functional size of the organism, and the development pattern, shape, and from of a newly forming animal. Embryology focuses on answering the question of how cell division and specialization is regulated so that eventually a fully functioning new organisms can develop. The environment affects developing embryos because they are extremely sensitive to the conditions that surround them. This experiment is observing the embryo development of Zebrafish when exposed to the tracer dye called Rhodamine. The observation of Zebrafish embryo development was not chosen at random, the reputation of this small tropical fish was firmly established during the nineties when big genetic screening projects led to the discovery of hundreds of mutants that gave insights into the functions of essential human developmental genes. These experiments and studies that proved to be very informative used Zebrafish for mainly two reasons, one being the optical transparency of the Zebrafish embryos.This makes observation very simple and also the use of fluorescent markers can be used to ‘light up’ different cells and organs in the zebrafish embryo. The other reasons is the Zebrafish reproduce quickly and large amounts. This gives a large sample for studying. Lastly, Zebrafish provided useful information for experiments because the invertebrate embryos show how human embryos would behave in similar conditions. The hypothesis of this experiment is that the exposure of the tracer dye, Rhodamine, will not affect the growth rate, development structure or function of the embryo. The Rhodamine will simply dye the transparent embryos, providing a better view of the newly forming animal. For this experiment, the zebrafish embryos were approximately 18 to 36 hours old fertilized eggs at the beginning of observation. The embryos hatch from translucent egg cases and come larvae at three to four days after fertilization. Materials and Methods:
To begin, a tub of Zebrafish were delivered to the lab the day before the lab. The embryos developmental stage was observed on day one. Next, the control group was set up by adding 20 mLs of Deerpark water and 20 healthy translucent embryos. The embryos were transferred by using a dropper, therefore, there was extra water added. In a separate petri dish, the same steps were taken, but there was 19mL of Rhodamine added as a tracer.
The next steps were done to both the experiment and the control groups. Over a span of two weeks, the embryos were observed for the following: number of dead eggs, living embryos in egg case, hatchings, dead hatchings, stage, and heartbeat detection. On day five, 10 mLs of water was removed, along with any dead eggs due to the mold that grow. Next, 25 mLs were added to the petri dish with living fish. All dead emtryos were saved in paraformaldehyde. On day seven, 5 mLs of water was removed, along with any dead hatchings. 10 mLs of water and two drops of paramecium were added to the group. On day 4, 7, and 11 the embryos were observed with a compound microscope. A few drops of the sample with the organism was added to a depression slide and viewed at the 4x objective. On 03/06 the embryo’s measured an average length of the fin to be 20 um, the eye measured an average of 13 and the diameter of the eye was 8. There was a visible heartbeat. It was also noted that the older the fish grew, the slower they swam.
All of the information was found on pages 60-61 of the General Biology 2 Laboratory Manual written by Meg Bentley, Kathryn Walters, and Nancy Zeller.
Conclusion:
My group had a very rough time observing this data. The weather delaying class made it difficult for my partner and I to do the observing together. Anther problem that we ran into was the fact that the petri dishes containing the experiment and control embryos were spilt. It was obvious the water just didn't evaporate, since there were no fish or water in the dish. The experiment was completely empty after only two days of observation. After the experiment went missing, only the control was observed. Those results are conclusive, however we have nothing to compare them to.
This image shows the developed Zebrafish on 03.06.15 in the control sample. File:IMG 6184
CL
Determining Species of Bacteria using the 16s rRNA Gene 03.05.2015
Purpose: The purpose of this experiment was to find specific bacteria from the Hay Infusion Culture to help determine the bacteria that is a part of the National Wildlife Habitat transect observed and how they relate to the organisms already determine. The PCR was used because this amplifies the 16s rRNA gene that is used in determining the name of the bacteria. Different bacteria have unique enzymes with specific metabolic activity, resulting in different DNA. Thus, the PCR using primer sequences specific for these genes can be used to identify the species (Bentley 27). These sequences will also help distinguish is these species are related, if so, how closely they are related which will in turn show the great biodiversity present. The isolation of the DNA from the bacteria in the colonies with the use of primers and PCR to selectively amplify the 16s rRNA gene, which is diverse and specific to each species (Bentley 32)
Materials and Methods: There were four bacteria characterized from the Hay Infusion Culture created with materials from the National Wildlife Habitat Transect. The four plates consisted of two nutrient agar plates and two agar plates containing tetracycline. With specific instructions from the Lab Manual, the steps described below were followed.
To begin, a single colony of bacteria was put into a small test tube, containing 100 ul of water. The tube was then heated at 100 degrees Celsius, incubating for approximately 10 minutes. The colony, still in the tube, was centrifuged for five minutes at 13,400 rpm with a 20 ul mixture of water and primer added. The tube was then shaken to mix the ingredients. Lastly before being placed in the PCR machine, 5 ul of the supernatant was transferred to the 16s PCR. The PCR products were ran under an agarose gel. Next, the DNA was purified for sequencing. The amplification of the rRNA provided the specific sequence needed to identify the species of bacteria. This actual name of the bacteria was found through BLAST.
https://clims3.genewiz.com/links.aspx?oId=4p6LZ4Isjwk=&ref=00 The link above will show the sequencing reactions which will be needed to begin the BLAST process. First, the sequence must be matched with a corresponding MB# tube label. Once the sequence is found, select to “View” the SEQ File associated with the tube. This will bring up a new window with the sequence of the 16s PCR product. The sequence will then be copy and pasted into this lab notebook. Now the BLAST process can begin. Copy and paste the sequence in the “nucleotide blast”. A graphic summary will appear, with descriptions of matches. Using the highest match, it can be confirmed that the colony morphology, gram stain and bacterial cell shape are correct.
All information was taken from pages 25-33 in the General Biology 2 Laboratory Manual, written by Bentley, Walters-Conte and Zeller. Also, information was taken from the TA notebook on OpenWetWare, written by Laurie Stepanek. Results:
Raw sequence of sample 1 (Nutrient 10-3):
NNNNNNNNNNNNGNAGGCTANNNTGCAGTCNANCGGTNGTANNNCNNTANNGNATTCNTGANANCGCCGTAGAGGGGCGAATCATGNCTGCNACCCGGCCTGGTCGTGGGGATNNCCTTTCCAAAGGAANATCTNACCCCNTAACGTCCNANNGNANAAANTTGNGACTTCACTCCTTTGGATATCGANGGGCCNCGTCGGANNNNNTNNTGNNAGGGNNNTGGCCCACCNANGNATGNNCTTTAGGGGGNCGANAGGGTGATCCCCCCACTGNACNGAGCANNNNCNNACTCCNACNGNAGGCAGCANNGAGGANNATTGNACNATGGGTGAGAGCCTGATCCNCCATGCCGNNTGNNNGATGANNGCCCTATGGGTTGTAAACTTCTTTTGATNNGGATAAACCTTTCCACGTGTGGAAAGCTGAANGTACTATACGAATAAGCACCGCCAACTCCGTGCCNGCAGCCGCGGTAATACNGANGGTGCANNCGTTATCCGNATTTATTGTATTTAAAGNATCCNTANGCGGATCTGTAANTCAGTGTTGAAATCTCACNNNTTAACTGTNAAACTGNAATTNATACTGCAAGTCGNGACTAAAGTATANNTGACTNGAATAAAAANTGTAGCAATGAGTACAAANATATTACTTAGTACNNAAATTGCGAGNGCAGGTGGATGNCTCTCTGNNTGNNACCNACGAGA
Raw sequence of sample 2 (Nutrient 10-5):
NNNNNNNNNNNNNCNANNNTGCAGCCGAGCGGTAGAGATTCTTCGGAATCTTGAGAGCGGCGTACGGGTGCGGAACACGTGTGCAACCTGCCTTTATCAGGGGGATAGCCTTTCGAAAGGAAGATTAATACCCCATAATATATTAAGTGGCATCACTTGATATTGAAAACTCCGGTGGATAAAGATGGGCACGCGCAAGATTAGATAGTTGGTAGGGTAACGGCCTACCAAGTCAATGATCTTTAGGGGGCCTGAGAGGGTGATCCCCCACACTGGTACTGAGACACGGACCAGACTCCTACGGGAGGCAGCAGTGAGGAATATTGGACAATGGGTGAGAGCCTGATCCAGCCATCCCGCGTGAAGGATGACGGCCCTATGGGTTGTAAACTTCTTTTGTATAGGGATAAACCTTTCCACGTGTGGAAAGCTGAAGGTACTATACGAATAAGCACCGGCTAACTCCGTGCCAGCAGCCGCGGTAATACGGAGGGTGCAAGCGTTATCCGGATTTATTGGGTTTAAAGGGTCCGTAGGCGGATCTGTAAGTCAGTGGTGAAATCTCACAGCTTAACTGTGAAACTGCCATTGATACTGCAGGTCTTGAGTAAGGTAGAAGTAGCTGGAATAANTANTGTANCNNTGAAATGCATAGATATTACTTANAACACCAATTGCGAAGGCANGTTAGTATGTCTTAACTGACNCTGATGGACNANNNCGTGNGGAGCGAACANGATTGNATACCCTGGTNGTNNNCNCCATAAACGATGCTAACTCNTTTTTNGNCTTCNGCTTCAGANACTAANNNAAANTGANAAGNTTAGNNNCCTGGGAGTANGTTTCNCAGANTGATNAAANNAANTNACGGGNGCCCNCGCNCCCNCTNNAGTTATGTNNTTTATTNNTTNATACNGANANNNNNNNAGNNANNNNTAATGGCGANNGNGGGNGNNNGNNNNNGNGNGTGCNTTNNTCANNTTNGNNCTTNNTNCNTGGCNNNNGNGGGNNNGGTNNCCNNNNNNNNGNNNNNNGNNNNNNNNNCNGNNNGGNN
From the BLAST process, sample 1 was identified as Chryseobacterium sp. EECC-510. Sample 2 was identified as Chryseobacterium sp. THG A18.
Conclusion
Both of the bacterial samples identified as a part of the genus Chryseobacterium. From data observations made on the bacterial sample in the lab, neither samples would have been identified as Chryseobacterium. Sample 1 was observed to be gram negative like Chryseobacterium but the bacterial cells were identified as round not rod shaped. Similarly, sample 2 as observed to be gram positive unlike the Chryseobacterium, but did have the same rod shaped bacterial cells. The colonies of the two samples were observed as a deep bright yellow similar to the description common to Chryseobacterium. There could be an error in the data observed in lab. There should be more samples observed and grown before any conclusions can be made about the data not matching up. Also, there could have been an error in any of the processes, considering I don’t have much experience with gram stains. This could be a factor in the data found.
CL
Biodiversity of Vertebrates in The Wildlife Habitat 02.18.15
The purpose of this report is to determine the vertebrate organism that might inhabit the National Wildlife Habitat transect. There have many protist, fungi, invertebrates and plants identified in the transect. The National Wildlife Habitat transect was observed during the months of January-Feburary. The average temperature during this time was between 15 degrees and 40 degrees F. The transect was obsessed for about 20 minutes in the evening, around 4:00 pm.
Five different vertebrate organisms that were observed to have inhabit this transect include: squirrel, mouse, rabbit, Charadrius vociferus (bird), and the Setophaga pinus (bird). They all seemed to be looking for food or prey. The transect is very open, there isn't many places for the animals to find shelter at this time of the year since the ground is frozen and all of the leaves have fallen.
The invertebrates and the vertebrates have a very symbiotic relationship; both benefit from each other, as well as the plants, protist, bacteria. They all provide a balance niche for each other to live in.
CEL
Biodiversity of Invertebrates in The Wildlife Habitat 02.18.15
The purpose of lab five was to show the immense biodiversity of invertebrates in leaf litter. Although they can’t be seen with the bare eye, they still play a huge roles in the ecosystem. In this lab, five different invertebrates were identified in the leaf litter. Also, three different types of worms were observed and their different types of movements were compared.
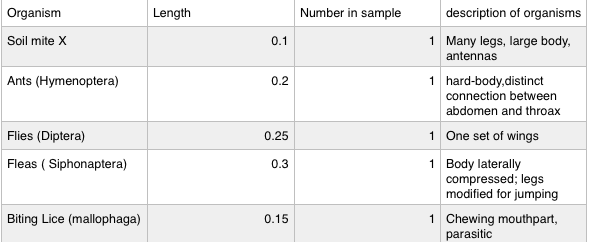
Each invertebrate was only identified once in the culture. The average size was 0.2. The smallest invertebrate found was the soil mite X. The largest invertebrate found was the fleas.
In procedure one, the acoelomate, planaria, was observed with the dissecting microscope. The planaria moves by a gliding motion, using muscular contractions. The planaria move by their cilia. The nematodes were also observed, however a compound microscope was used. The nematodes have longitudinal muscles, making them move in a hectic like fashion. There bodies move in a sin like movement, which is not a very efficient way to move. To change direction, it has to coil its body into almost 360 degrees and then whip its self back the other direction. Lastly, the coelomate, Annelida, was observed. Annelida move by the muscles and setae on the stomaches. The setae are used to help the organisms slow down. There are muscles in circles around the body and also muscles running the length of the body. The circular muscles squeeze the front end forward and the long muscles squeeze together the rear, moving the body toward the front end.
CL
Identifying Plantae and Fungi in the Wildlife Habit Transect: Lab 4 02.15.15
In the assigned transect, National Wildlife, there is an array of plants present. The plants are all characterized for three general feature: presence of Vascularization, presence of Specialized Structures, and mechanism of Reproduction.
A plastic bag was taken to the transect to collect 500 grams leaf litter and soft soil. This was used to set up the Berlese funnel, which is used for collecting invertebrates in the next lab. There were five samples taken: long leaves, moss, back bush, tree, and front bush. The green, long leaves were taken from one singular tree, measuring to be about a foot long. The moss measured to be approximately a foot in diameter and less than an inch in height. It was light green colored, which could be due to the cold weather. The sample taken from the back bush was a very dark green and measured to be approximately one foot in height. The tree sample was taken from a tree in the middle of he transect that measures to be approximately eight feet in height. The leaves were a very light green, and very small in size. The last sample was from a front bush that was approximately half a foot tall with purple/greenish leaves. All of the samples taken were vascular plants, except the moss sample. The long leaves, sample from the front bush, and the tree were characterized as monocot, meaning their structures resembled a leaf developed as a single structure. The sample taken from the back was determined as dicot, developing as pairs. The moss was neither because it does not have seeds or leaves. Fungi are decomposers that live in niches and are crucial for the survival of the biosphere. Their metabolisms release carbon dioxide into the atmosphere and nitrogenous materials into the soil and surface waters (Lab Manual pg. 42). Both of these products are used by plants to produce carbohydrates, which are then consumed by other animals. Non of the samples collected were in the fungi domain.
CL
Gram Stain Contrast With and Without Tetracycline 02.03.15 There are many ways for identifying and studying bacteria. Scientists use many different staining techniques to observe unicellular organisms via microscope. The most common staining technique is the Gram stain, showing with microorganisms are gram positive and gram negative. They also use morphology to classify microorganisms. There are three early classifications of basic bacterial shapes: bacillus, coccus, and spirillum. Microorganisms are also classified by their nutritional essentials. Different bacteria have unique enzymes encoded with different DNA sequences for metabolic activity. This DNA sequence can also show how closely related each microorganism is to each other. In the agar plates inoculated from the Hay Infusion Culture, there were visible colonies and fungi that had grown over the past week. The plates with the antibiotic had a substantially lower amount of colonies growing, as opposed to the plates without the antibiotics. The higher the dilution, the less colonies survived. Bacteria that did grow in the plate with the antibiotics may be insensitive to the antibiotic and even antibiotic-resistant. To begin the Gram Stain procedure, a loop was sterilized over a flame and then cooled. A tiny amount of the surface of the agar was scraped and added to a slide with a drop of water. This was done with plate. The slides were then heated until the water dried. This took different lengths of times depending on how much water was added to the slide. The slides were placed on a staining tray, then covered with crystal violet for one minute. The crystal violet was washed off and then the bacterial smear was covered with Gram’s iodine mordant for one minute and then rinsed with water. The bacteria smear was then decolorized with 95% alcohol for 20 seconds. Then it was covered with a safranin stain for 20-30 seconds and then rinsed with water. All slides were then left to air dry and then examined under a compound microscope. This was used to identify the bacteria as gram positive or gram negative. Only four colonies were observed: 10^-3, 10^-5 (both without tetracycline) and 10^-3,10^-5(both with tetracycline). The agar plates with the tetracycline had a substantially smaller amount of colonies. They were very spread out. The agar plates without the tetracycline were very clustered with very small colonies. It was very evident in the data that the tetracycline did reduce the amount of bacteria produce, however it did not eliminate it completely. This shows that the bacteria is either insensitive to the antibiotic or is antibiotic-resistant. I believe this is why scientists are starting to use organic compounds as antibiotics. According to an article published in Janurary, these organic compounds have proven to be less resistant and more effective over time. Using tetracycline is a great example of bacteria becoming resistant to antibiotics. Overtime, tetracycline would become ineffective.
CL
01.29.28 Analyzing Specimen Found in Hay Infusion Culture
In lab two, the Hay Infusion Culture that was grown in lab one was examined and multiple algae and protists were identified. There looked to be more specimen growing on the top of the ecosystem, therefor I predicted that there would be a more diverse finding of algae and protists found in the top of the ecosystem. The purpose of this experiment was to determine different organisms by characterizing their appearance, motile abilities, functions, and whether they are photosynthesizing, or non-photosynthesizing.
There were many materials used to observe the algae and protist found in the ecosystem. The samples were extracted from the Hay Infusion Culture using a pipette and then placed on a slide. The samples were viewed using a compound microscope under the 40x objective. To identify the organisms found in the sample from the Hay Infusion Culture, a dichotomous key titled “Ward’s Free-Living Protozoa” was used in reference. This key showed characteristics, names, functions and pictures of the algae and protist found, making the identification process easier.
There were two organisms identified from samples taken from the top of the Hay Infusion Culture. There were many different plant lives growing on the top. The first organism identified was an elongated, greenish figure with one flagella. This fit all of the characteristics and image of the Eugiena from the dichotomous key. The second organism identified from the top of the culture was a small, oval shaped body with many flagella. According to the description on the key, this organism is called Colpidium SP. There were three organism identified in sample taken from the bottom of the Hay Infusion Culture. The first one was a small, green, oval body with a single flagella known by the name Chlamydomanas. The second organism that was identified was a creeping, white organism known by the name Arcella. The last one identified in the sample taken from the top was also found in the bottom. The organism that was found in both niches in the ecosystem was Colpidium.
The ecosystem being contained in the jar had an earthly smell with a green/brown layer growing on the top. There was a substantial amount of water gone from the original amount measured when the culture was created. The ecosystem resembled a swamp. The purpose of observing this ecosystem was to identify the vast differences within small niches in a large ecosystem. By observing different protists and algae using a dichotomous key, many different organisms in the ecosystem were identified. This provided evidence that niches within an ecosystem can be vastly different.
CL
01/26/15 Creating the Hay Infusion Culture
Purpose and background My group member and I are analyzing a transect on AU’s campus. My partner and I were assigned to transect number two, the Certified Wildlife Habitat. We have taken time to observe the ecosystem, listing the abiotic and biotic objects. We also collected samples from the transect with interest in identifying the protist, bacteria, plants, invertebrates and animals in the transect.
There were many materials used in lab one. The sample collected from the transect was held in a covered tube. This sample was then transferred into a Hay Infusion Culture. This culture included: 11.8 grams of soil/ground vegetation sample collected from the assigned transect, 500 mLs of deer park water, 0.1 gm of dried milk. These ingredients were contained in a medium sized jar, with an open lid. The Culture was left for a week before being observed.
The assigned transect was a very open space, with only four trees. There were few bushes, but many small plants. The ground was completely frozen, making the sample difficult to collect. Through observing a transect, many bacteria, plants, and animas can be identified. This will help determine characteristics of each, and the environment that suits them the most.
CL