User:Anthony Salvagno/Notebook/Research/2010/05/11/Gel of Lambda DNA
From OpenWetWare
Jump to navigationJump to search
Quick Navigation: Pages by Category | List of All Pages | Protocols
Gel Results
Some notes:
- Yesterday's gel was a 0.8% but after some reading I decided to do a 0.5% gel. I hope it works out well.
- I am running the gel at a nice and low voltage of 60V. According to Molecular Biology Protocols, you get the best resolution between 1-5 V/cm and max decent resolution at 5-8 V/cm. I usually go way over this. But since I want good resolution and I have a very low gel concentration I will use less power.
- I did some extrapolation. I found the gelpilot manual and came across an interesting chart. I used the data and put it into excel to get the equation of the graph generated and then figured out what the point would be for a .5% gel. The equation is off a bit but it says about 6000bp. I will estimate around 8000bp for my purposes. Table is below.
| % Gel | bp marker |
|---|---|
| 2.5 | 700 |
| 2 | 1000 |
| 1.5 | 1800 |
| 1 | 3000 |
| .8 | 5000 |
Pics and Comments
Lane assignments:
- lambda DNA prior to reaction
- lambda DNA post Klenow (ie with fill-in)
- lambda DNA with neutravidin
- lambda DNA with qdots
-
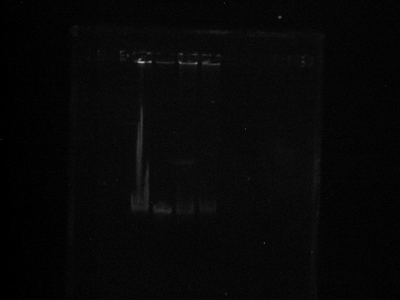
This is a picture after staining prior to destain, which I am doing now.
-
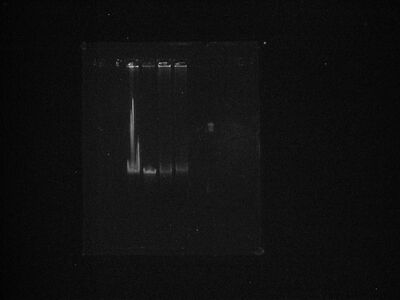
This is the destain picture in b&w with brightness adjustment.
-
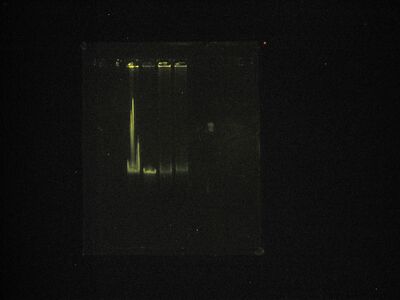
Destain in enhanced color.
-
Original destain.
Comments:
- Image 1:
- What the heck is up with that huge smear of gook in lane 1? Idk, but I saw something similar to that yesterday in my failed gel. It reminds me of E. coli genomic DNA. Search through my early notebook for what I'm talking about.
- It is tough to tell a shift between lanes 3 and (2 and 1), but I see a shift between lanes 4 and the rest. I guess that means the QDs stuck?
- The neutravidin lane (3) is smeared so I'm using that as proof that it worked.
- Of course I won't know for sure until I check it out in a microscope.
- Image 2
- No new insight, really.
- You can see the shift in lane 4 a little better here. Because the fragments are so large, I'm guessing any kind of noticeable shift is huge.
- My only guess as to the differences between lanes 1 and 2 is the Klenow working. Because the ends of lambda DNA are complimentary you can get concatamers and such, but the fill in will prevent that. The only problem with that theory is that we aren't seeing a banding going on in the first lane. Still I suppose there has to be some difference otherwise all 4 lanes would exhibit the same behavior.
- In Image 1 you can see a band above lanes 3/4 but in the redone image that band disappears. Artifact of photography I suppose.
- There was no color image for the first picture because I took the picture as a black and white.