User:Anthony Salvagno/Notebook/Research/2010/02/16/DNA Concentration Tethering Part 2
Quick Navigation: Pages by Category | List of All Pages | Protocols
This is started from yesterday.
Protocol
Results
-
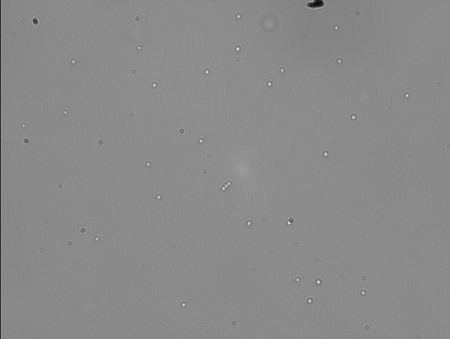
This sample has no DNA. I saw a couple of jigglers, but they were barely moving. I also saw a jibbling bead that broke free and diffused away. I don't know what to make of that.
-
This sample has 1:20 dilution of DNA in Popping Buffer. Only 4 beads in this FOV were stuck. The rest looked like good tether candidates. That's awesome because there are a lot of beads here.
-
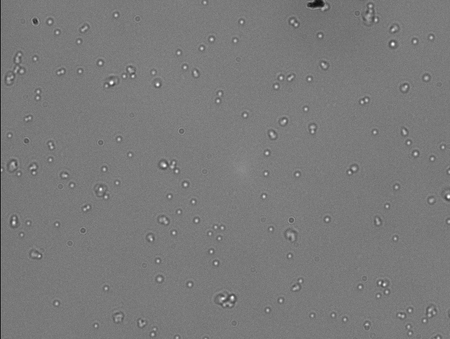
This sample has 1:50 dilution DNA in 1x POP. The ratio of jiggling:stuck was about 2:1 and so I am pleased. Just about every FOV had this ratio and only a few were super tight jigglers. A couple also were super floppy.
-
This is the 1:100 dilution. I am very pleased with this result because like the rest I had a high amount of jiggling beads compared to stuck beads. (About 2:1) This is getting down to the range that I will be tethering with after gel extraction.
I would say these experiments are a resounding success. Also I think I will tether with pBR322 to get a feel for what it is like to tether for unzipping. Let's compare these results to last Fri's experiment:
-
This is a typical FOV of the no DNA sample.
-
This is an acquired image from the sample with the most DNA (around 300ng/ul).
-
This is a view of the 1:5 dilution of DNA.
-
This sample is the 1:10 dilution.
Comparing the two sets of data I would trust Fri's less because the DNA isn't diluted with Popping buffer. It also seems I didn't do as good a job removing bead clumps from the first batch of experiments. I would say bead concentration wise, my latest results are in line with Fri's excluding the 1:10 dilution of DNA. That is to say the amount of beads per sample decreases each time except in 1:10 where it has even less beads (overall) than the 1:100 sample. Hmmm...