User:Anthony Salvagno/Notebook/Research/2010/02/12/DNA Concentration affects during Tethering
Quick Navigation: Pages by Category | List of All Pages | Protocols
Background
This is going to be a To be continued... kind of post because I can see doing this many times for some decent data. Anyways I think it is a great idea to see how the [DNA] affects tethering, bead sticking, etc. Yesterday I purified some PCR DNA from a reaction Brian did in December:
<html><iframe width="800" height="300" frameborder="1" scrolling="no" src="http://openwetware.org/wiki/User:Anthony_Salvagno/Notebook/Research/2010/02/11/Tethering_Experiments#Nanodrop_Result"></iframe></html>
This will be my starting concentration.
Setup
I will dilute the DNA several times in this fashion:
- Starting amount
- 1:5 dilution - 2ul start in 8ul water
- 1:10 - 1ul start in 9ul water
- 1:20 - 0.5ul start in 9.5ul water
- 1:50 - 1ul 1:5 in 9ul water
- 1:100 - 1ul 1:10 in 9ul water
- 1:500 - 1ul 1:50 in 9ul water
- 1:1000 - 1ul 1:100 in 9ul water
I don't think I will get to try all these dilutions today. But I will tethering with the most concentrated amounts and work my way down and continue next week.
Tethering
I sonicated the beads this time for 15 seconds. The splashing inside the tube was immediate as in as soon as the tube hit the liquid, there was splashing. After about 5 seconds there was significant splashing and I gave it an extra 10 seconds for good measure. I am not spinning it down this time in case the spinning allowed for clumps. I am also going to try my precleaned glass.
Normally I wash with BGB after each step and I use 50ul (5 sample volumes) to perform the wash. Here I used 10ul for one wash because I was running low. I am doing it after the DNA stage because I think that is where it is the least crucial to wash out of the 3 wash points (not that they aren't all important). I am doing 10ul instead of nothing because I think it is important to get some untethered DNA out especially in the high concentration sample.
Results
I'll be looking for number of stuck beads per field of view (FOV) and number of jigglers compared to number of stuck beads. I will also look at the jiggling range (more DNA means more tethers can attach to a single bead so beads could jiggle less at higher concentrations). I'll try and post images of each sample here.
-
This is a typical FOV of the no DNA sample. The image is a little blurry because of vibration in the camera, but in this particular image there are 3 stuck beads and 1 stuck clump. In the entire sample I saw NO jigglers. This particular image has one of the higher amounts of stuck beads as most FOV's had one or two stuck beads and nothing else.
-
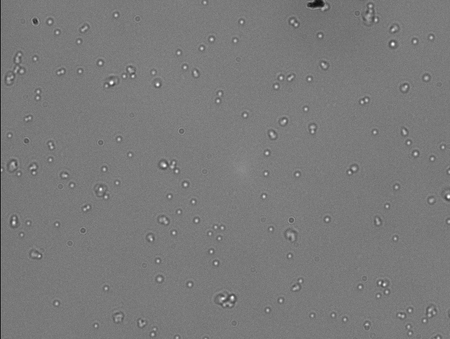
This is an acquired image from the sample with the most DNA (around 300ng/ul). As you can see there is a TON of stuck beads. I counted at least 20 jigglers in this image (I gave up after a while). I suppose a lot of these are multiple tethers as well because of the rest of the images. Every FOV looked just like this. I couldn't get a good approx of the Stuck:Jiggle ratio because of the high volume of beads but if I had to guess I'd say like 5:1.
-
This is a view of the 1:5 dilution of DNA. I don't know how reliable my analysis of this sample is because a bubble as wide as the sample chamber swept through when washing. Here there were more jigglers than stuck beads in almost every sample, but a lot of them were floppy jigglers. In this particular image there are no stuck beads. My analysis of stuck:jiggle is 1:2 (more jiggle than stuck), but again I don't trust it because of the bubble.
-
This sample is the 1:10 dilution. The number of stuck beads was easy to deal with because of the low amount of DNA. I don't remember what the stuck:jiggle ratio is, but it was near 1:1, maybe 2:1 at the most. There were also a lot of floppy tethers though. In this image most of the beads were very floppy tethers (with range of motion being on the order of 1-1.5um). But you can't tell that because you are looking at a picture!
- Steve Koch 22:25, 12 February 2010 (EST): Very nice notebook page and good experiments. Do you think the "floppy" tethers are the good ones? It does seem, based on your evidence, that the undiluted samples are too concentrated. Looks like 1:5 is a good bet.