User:Andrea Loehr
Contact Info
- Andrea Loehr
- Harvard-Smithsonian Center for Astrophysics
- Cambridge, MA, USA
- Email me through OpenWetWare
I work at the Harvard Smithsonian Center for Astrophysics. I learned about OpenWetWare through the Church Lab, and I've joined because I have started to work on a project with the Personal Genome Project (PGP). Our goal is to set up a volunteer computing project PersonalGenomes@Home, analogous to seti@home.
Building PersonalGenomes@Home
The Personal Genome Project (PGP) will publicly sequence the DNA of 100.000 volunteers and make the results freely available. The analysis of the raw data entails a huge computational effort and will require much computational power. The PGP is an open source project. In this spirit we would like the data analysis to become an open source community effort through distributed computing with 'PersonalGenomes@Home'. The part of the data analysis that will be initially distributed to the community is the analysis of raw Illumina images to produce reads and alignment against a reference. We are currently setting up the open source software packages Swift and BOINC as well as Tranche and Free Factories.
Running Swift
Swift is designed to process a single tile of Illumina data at a time. It can be run in two modes: 1) base-caller only (processes intensity files produced by the Solexa pipeline) and 2) image analysis. The output files are purity-filtered and non purity-filtered reads in fastq format, a run report, an intensity file and files listing all processed images.
Processing PGP Data
After verifying the functionality of the code with example data (available through Swift), we have run Swift on tile 87 of one PGP2 data set. Images are 7.1MB in size and the processing resources required exceed the abilities of my personal computer (1.5MB RAM). Swift is designed to process one full tile of Illumina data at a time. Attempts to process less than 36 cycles remain unsuccessful. Keeping in mind that the goal is to perform the data analysis on a 'regular' computer, we did not move to larger memory capacities. Instead, the large images were cropped into three horizontal thirds of 2.4MB each using ImageJ, an open source image processing tool. Images overlapped by 10 pixels to avoid errors caused by edge effects. The one tile was then individually processed in three sub-tiles using Swift in approximately 20 minutes.
Finding the MFN2_R364W Variant and Control
To prove that Swift produces correct results, the stack 87 data were searched for the MFN2_R364W variant and stack 27 for the control. The last numbers are the x and y coordinates on the images. Differences in aligning techniques between the Illumina pipeline and Swift result in minor discrepancies in coordinate assignment.
Previous analysis had shown a T in the 11th cycle:
HWI-EAS0184_1:2:87:783:1097
GCACACGGTCTGGGCCAaGCAGATTGCAGAGGCGGg
The Swift analysis confirms this result:
@mid:784:1097
GCACACGGTCTGGGCCAAGCAGATTGCAGAGGCGGG
Analogous, the control sequence was extracted from stack 27.
Previous analysis had shown a C in the 13th cycle:
HWI-EAS0184_1:2:27:936:1555
CAGCACACGGTCCGGGCCAAGCAGATTGCAGAGGCG
The Swift analysis confirms this result:
@control_bot:935:1555
CAGCACACGGTCCGGGCCAAGCAGATTGCAGAGGCG
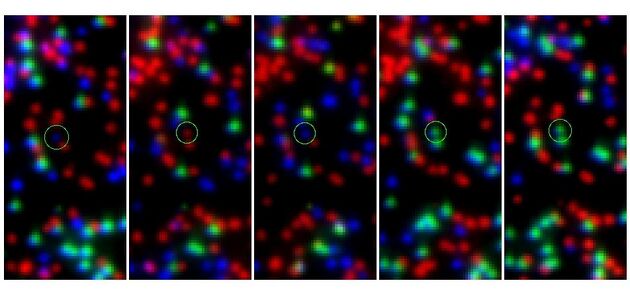
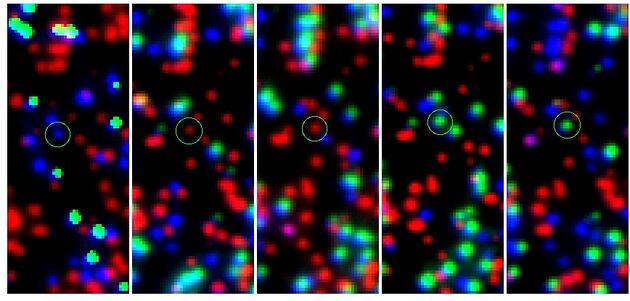
The results are illustrated in Figures 1 and 2. Images from the three cycles C, G, and T were loaded into the three
color channels red, green, and blue of SAOImage ds9 to create a combined
three-color image. The images show 50x100 pixels centered on the location of the
variant and control sequence as determined by Swift. Fig. 1 shows cycles 9 through 13 of stack 87 for the variant MFN_R364W. Colors are red: C, green: G, blue: T. The expected sequence is TCTGG (blue, red, blue, green, green). The location of the variant is circled.
Fig. 2 shows cycles 11 through 15 of stack 27 for the control sequence for the variant MFN_R364W. Colors are red: C, green: G, blue: T. The expected sequence is TCCGG (blue, red, red, green, green). The location of the variant is circled.
Next Steps
Next, we will automate the image processing with a shell script and ImageMagick. We will install and run the Swift software on BOINC.
Comments and suggestions are welcome.
Education
- 2003, PhD Physics, Rheinische Friedrich-Wilhemls Universitaet zu Bonn, Germany
Research interests
- Dark Energy, Cosmology
- Whole Genome Sequencing
Publications
- Staniszewsky et al., 2008, 'Galaxy clusters discovered with a Sunyaev-Zel'dovich effect survey' arXiv:0810.1578