The BioBricks Foundation:Standards/Technical/E.coli promoter standard
Jason R. Kelly 02:11, 30 March 2008 (EDT):Seems like we should just make a decision about where to locate transcription start site (+1 site) in E.coli sigma70 BB promoters. There's excellent previous discussion on the topic here (I also copied a portion of it below).
A proposal
- Jason R. Kelly 05:36, 30 March 2008 (EDT): Option A -
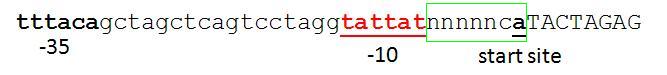
This basically follows from Chris & Reshma's discussion below. The only addition is that the standard requires a defined spacing between the -10 box and the CAT sequence. The A in the CAT sequence should be the transcription start site. The reasonable spacing along with the use of CAT (which is the 'consensus' -1,+1,+2 sequence - see table) should hopefully lead to predictable transcription start at the 'A'. Unfortunately the current Berkeley promoter library / R0040 don't conform to this standard. I suspect their transcriptional start is at the first A in the BB junction, but it's hard to know because the sequence isn't obviously optimal for a transcription start though it's not bad (see fig).
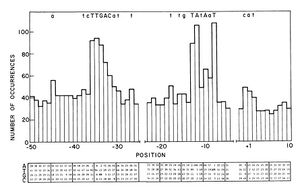

- Jason R. Kelly 04:59, 30 March 2008 (EDT):The other option is to adopt the R0040/Berkeley promoter set as the promoter standard. E.g. (-10box)nnnnnc(BBjunction). Downside here is that (1) I don't know how dependent the "non-consensus" transcriptional start will be on the n's (so might get different start sites w/ diff promoters) and (2) I don't know exactly where the transcriptional start is in the first place. Though I might just not be up to speed on best way to predict start site (ref below was best I could find). Of course this standard would have the advantage that we already have some parts in the format ;)
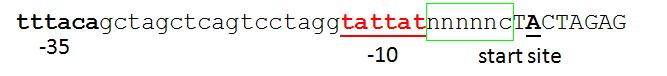
Continued Discussion
Joey Davis 07:30, 31 March 2008 (EDT):
I've added a few thoughts about standard promoters::
I wholeheartedly agree with the need to standardize promoter construction and decided to throw in a few other thoughts. In particular, I think we should focus on creating a good standard for the interface between a promoter and an RBS – I mention it briefly below.
1. Focus on one family of sigma factors:
I would recommend focusing on the sigma70 family as this encompasses most of the factors we commonly think about (RpoD,S,H,E). Further, the sigma54 family is commonly activated by remote elements which would either make for very large promoters or would greatly limit composability.
2. Functional organization of the promoter:

Naively, promoter strength is a function of 3 variables: holoenzyme affinity for promoter, equilibrium between closed and open complex and efficiency/frequency of promoter escape (this includes all events between opening of DNA helix and clearance of the promoter – in particular initiating synthesis of the first phosphodiester bond, idling/stuttering leading to the generation of nonproductive RNA oligos and finally clearance of the promoter). In the absence of activation/repression, the -40 to +15 region controls all 3 of these variables (-35,-10 define binding, -10-+4 define DNA opening, +1-+20 define promoter escape) – I think this is the absolute minimum we should use to define a promoter. More realistically, we should probably include back to -100 and forward to +20 as the vast majority of both repressors and activators act in this region (see a sampling of ~200 coli promoters below).
- Joey Davis 21:44, 31 March 2008 (EDT): Moreover, the "up" region which binds alpha subunit sits around -40 - -60 making this region all the more important to include.
It seems using this larger definition would be particularly useful when composing parts – you wouldn’t have to worry about an unforeseen repressor binding site which is coincidentally just up- or down-stream.
- Collado-Vides J, Magasanik B, and Gralla JD. Control site location and transcriptional regulation in Escherichia coli. Microbiol Rev. 1991 Sep;55(3):371-94. DOI:10.1128/mr.55.3.371-394.1991 |
- Pérez-Rueda E, Gralla JD, and Collado-Vides J. Genomic position analyses and the transcription machinery. J Mol Biol. 1998 Jan 16;275(2):165-70. DOI:10.1006/jmbi.1997.1465 |
- Estrem ST, Gaal T, Ross W, and Gourse RL. Identification of an UP element consensus sequence for bacterial promoters. Proc Natl Acad Sci U S A. 1998 Aug 18;95(17):9761-6. DOI:10.1073/pnas.95.17.9761 |
3. Transcript stability
The promoter definition itself can have no influence on 3’ dependent or internal cleavage dependent mRNA stability – so that sucks – when we put different parts downstream of a given promoter, we will get different absolute steady-state levels of mRNA. However, ideally, the rank order (in terms of strength) of different promoters should be maintained. While we could include a standardized 5’ UTR in every promoter definition, I think (although with no profound justification) that this would limit promoter design too drastically (so much regulation and strength determination occurs downstream of the start site). Thus, if +20 were included in the promoter definition, the 5’ stability of the transcript would be an inherent characteristic of a given promoter and not of the downstream transcript it is driving (admittedly a bit odd).
- Jason R. Kelly 21:44, 31 March 2008 (EDT): So to be clear this wouldn't constrain the promoter sequence up to +20. (e.g. you need to define the promoter up to the +20 position, but there are no sequence requirements for positions +3 through +20 -- position +1 and +2 are A,T respectively.
- Promoter escape module from EcoSal - review covering why promoter should run through the +20 site.
- Jason R. Kelly 13:51, 4 April 2008 (EDT):I wonder how much a problem the RNA engineering community would have with giving up control of the first 20bp of the mRNA to the promoter. For instance stuff like a 5' stem-loop can stabilize mRNA. [5]
4. Transcription start site
I like Jason’s proposal – in particular I don’t think that enforcing a standard length between the -10 and the start or a defined CAT start is too limiting (some people decrease promoter strength using this region but I think that there are sufficient alternatives to justify always knowing the initiating position). It might be worthwhile however requesting something of the form TATTATnnnnBCAT (where B is C, G or T) as an adenosine 5 nt from the -10 can be used (albeit infrequently) to initiate.
5. Composition between promoters and RBS
Because folding of a 5’UTR can profoundly influence the apparent ribozyme binding/translation efficiency it seems we should provide some insulation between these components. One could imagine placing standardized insulation at either the 3’ end of the promoter part or alternatively at the 5’ end of the RBS part or both. Maybe someone with a background in RNA folding could weigh in on amount of insulation required or if this is even feasible. I guess the biggest concern is that you would get unexpected base-pairing interactions formed between the “RBS part” and the “promoter part” on the completed transcript. Because the loops in stem-loops can be so large I don’t see any easy way to fix this…ideas???
JCAnderson: The site: http://parts.mit.edu/registry/index.php/Help:BioBrick_Prefix_and_Suffix under the BioBrick Prefix section has a really critical piece of information on how to design biobrick basic parts, and I think we should add to that a preferred way of biobricking the promoter initiation site relative to the polylinker to avoid heterogeneity 5' to the biobrick junction. Again, it is an arbitrary standard, and the options are (with the transcription start in bold): Define it like r0040(and what iGEM2006 did for the family of constitutive promoters):
- ...ctACTAGT
Or have it in the biobrick site explicitly, something like:
- ...ACTAGT
So that nothing has to be re-made, and so that more native promoter sequence can be present in the part I lean towards defining the standard as the r0040-compatible version.
Clearly not all promoters are going to be compatible with this standard. Some promoters have operators that overlap or extend beyond the transcriptional start. When making basic promoter parts, one has to currently make an arbitrary decision as to where to put the 3' end of the promoter. It would be preferrable to have a standard.
- Reshma 11:12, 21 August 2006 (EDT): I agree that we should have a default standard for the promoter-RBS junction. But in looking at the sequence logo for E. coli promoters, I think the typical nucleotides for the -1 and +1 positions are CA. In the absence of any strong reason to go with another scheme, why not go with E. coli promoter consensus? So perhaps something like ...
- ...caTACTAGAG
- i.e. Pretty similar to the R0040-compatible version but with the transcription start site being a defined nucleotide where possible along with the nucleotide before. It might make it a bit more likely that the transcription start site occurs where we think it should occur.
Reference
- Hawley DK and McClure WR. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237-55. DOI:10.1093/nar/11.8.2237 |
- Emory SA, Bouvet P, and Belasco JG. A 5'-terminal stem-loop structure can stabilize mRNA in Escherichia coli. Genes Dev. 1992 Jan;6(1):135-48. DOI:10.1101/gad.6.1.135 |
- Horwitz MS and Loeb LA. DNA sequences of random origin as probes of Escherichia coli promoter architecture. J Biol Chem. 1988 Oct 15;263(29):14724-31.
- Promoter escape module from EcoSal - review covering why promoter should run through the +20 site.
Promoter design references
- Mayo AE, Setty Y, Shavit S, Zaslaver A, and Alon U. Plasticity of the cis-regulatory input function of a gene. PLoS Biol. 2006 Apr;4(4):e45. DOI:10.1371/journal.pbio.0040045 |
- Cox RS 3rd, Surette MG, and Elowitz MB. Programming gene expression with combinatorial promoters. Mol Syst Biol. 2007;3:145. DOI:10.1038/msb4100187 |
- Alper H, Fischer C, Nevoigt E, and Stephanopoulos G. Tuning genetic control through promoter engineering. Proc Natl Acad Sci U S A. 2005 Sep 6;102(36):12678-83. DOI:10.1073/pnas.0504604102 |
- Miksch G, Bettenworth F, Friehs K, Flaschel E, Saalbach A, Twellmann T, and Nattkemper TW. Libraries of synthetic stationary-phase and stress promoters as a tool for fine-tuning of expression of recombinant proteins in Escherichia coli. J Biotechnol. 2005 Oct 17;120(1):25-37. DOI:10.1016/j.jbiotec.2005.04.027 |
- Shimada T, Makinoshima H, Ogawa Y, Miki T, Maeda M, and Ishihama A. Classification and strength measurement of stationary-phase promoters by use of a newly developed promoter cloning vector. J Bacteriol. 2004 Nov;186(21):7112-22. DOI:10.1128/JB.186.21.7112-7122.2004 |
- De Mey M, Maertens J, Lequeux GJ, Soetaert WK, and Vandamme EJ. Construction and model-based analysis of a promoter library for E. coli: an indispensable tool for metabolic engineering. BMC Biotechnol. 2007 Jun 18;7:34. DOI:10.1186/1472-6750-7-34 |
- Murphy KF, Balázsi G, and Collins JJ. Combinatorial promoter design for engineering noisy gene expression. Proc Natl Acad Sci U S A. 2007 Jul 31;104(31):12726-31. DOI:10.1073/pnas.0608451104 |
Transcriptional start site references
- Hershberg R, Bejerano G, Santos-Zavaleta A, and Margalit H. PromEC: An updated database of Escherichia coli mRNA promoters with experimentally identified transcriptional start sites. Nucleic Acids Res. 2001 Jan 1;29(1):277. DOI:10.1093/nar/29.1.277 |
- Gordon L, Chervonenkis AY, Gammerman AJ, Shahmuradov IA, and Solovyev VV. Sequence alignment kernel for recognition of promoter regions. Bioinformatics. 2003 Oct 12;19(15):1964-71. DOI:10.1093/bioinformatics/btg265 |
- Gordon JJ, Towsey MW, Hogan JM, Mathews SA, and Timms P. Improved prediction of bacterial transcription start sites. Bioinformatics. 2006 Jan 15;22(2):142-8. DOI:10.1093/bioinformatics/bti771 |
- Web tool applying Solovyev method
- download of software tool applying Timms method
- list of web tools for promoter prediction
Repression references
http://jb.asm.org/cgi/content/full/181/10/2987
Activation references
- Ross W, Schneider DA, Paul BJ, Mertens A, and Gourse RL. An intersubunit contact stimulating transcription initiation by E coli RNA polymerase: interaction of the alpha C-terminal domain and sigma region 4. Genes Dev. 2003 May 15;17(10):1293-307. DOI:10.1101/gad.1079403 |
http://arjournals.annualreviews.org/doi/pdf/10.1146/annurev.ge.18.120184.001133?cookieSet=1
Initiation references
- Leibman M and Hochschild A. A sigma-core interaction of the RNA polymerase holoenzyme that enhances promoter escape. EMBO J. 2007 Mar 21;26(6):1579-90. DOI:10.1038/sj.emboj.7601612 |
http://arjournals.annualreviews.org/doi/pdf/10.1146/annurev.ge.19.120185.002035 http://www.ias.ac.in/jarch/jbiosci/18/13-25.pdf http://www.castu.tsinghua.edu.cn/course/ref/section-3/McClure85.pdf
Measuring Kit references
- Lu C, Bentley WE, and Rao G. A high-throughput approach to promoter study using green fluorescent protein. Biotechnol Prog. 2004 Nov-Dec;20(6):1634-40. DOI:10.1021/bp049751l |
Ponderings
- Jhdavis 20:37, 14 May 2008 (EDT):
1: Should we really rely on the the sigma70 type promoters or would it be better to pursue an orthogonal promoter system (maybe a polymerase/sigma system from another organism) such that we don't deplete the pool of coli pols.
- Jhdavis 20:37, 14 May 2008 (EDT):
2: Should the measuring kit have a second internal standard (like RFP under a known promoter) - this might help with things like copy number variations between different promoters, saturation of some transcriptional/translational component?
Notes from the MIT SB lunch
- Consider putting a terminator upstream of every promoter to ensure there is no trascriptional read-through.
- make a sub-set of promoters that have a defined trascriptional start and then 20 defined basis afterwards (phomoters), these would ensure identical mRNA from any phomoter put upstream of the same coding region.