T7.1
From OpenWetWare
Jump to navigationJump to search
| Project pages on Rebuilding T7 | |
|
back to Endy Lab | |
Background
Wild-type T7 is a superb organism for discovering the primary components of a natural biological system. However, our experience indicates that the original T7 isolate is not best suited for understanding how all the parts of the phage are arganized to encode a functioning whole. We decided to engineer a surrogate genome, which we designated T7.1, that would be easier to study and extend.
Goals
- We wanted to insulate and enable independent manipulation of all identified genetic elements.
- We wanted the T7.1 genome to encode a viable bacteriophage; at the start of this work, we were uncertain how many simultaneous changes the wild-type genome could tolerate.
Method
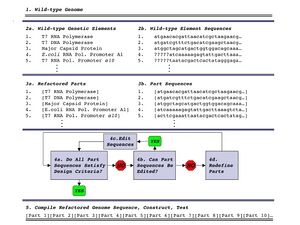
- Reannotation of the wild-type T7 genome, thus defining the functional genetic elements
- Specification of T7.1 genome design and sequence
- Construct sections individually
- Construct chimeric phages that contain replace a single wild-type section with a rebuit section
- Combine sections of rebuilt phage into a single rebuilt phage
- Characterize chimeric phage
Progress
- Reannotation of the T7 Genome -- The wild-type T7 genome is a 39,937 base pair linear double-stranded DNA molecule. We annotated the genome by specifying the boundaries of the following functional genetic elements: 57 open reading frames, 57 putative RBSs encoding 60 proteins, and 51 regulatory elements controlling phage gene expression, DNA replication, and genome packaging. A genbank file of the reannotation can be found here.
- Specification of T7.1 genome -- The designed sequence of T7.1 can be found here.
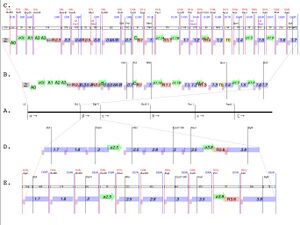
T7.1 genome design (sections alpha and beta). - Contructing the T7.1 Genome -- We constructed sections alpha and beta; the as built sequences can be found here
- Evolution of the T7.1 Genome -- We are starting to evolve the T7.1 genome to regain some fitness lost during the refactoring.
References
Refactoring bacteriophage T7
Nature/EMBO Molecular Systems Biology 13 September 2005 DOI:10.1038/msb4100025
Leon Y. Chan, Sriram Kosuri and Drew Endy
URL
PDF reprint
News & Views
October 2004 version