BME100 f2014:Group31 L4: Difference between revisions
| Line 50: | Line 50: | ||
| G31 - || Negative control || none | | G31 - || Negative control || none | ||
|- | |- | ||
| G31 1-1 || Patient 1, replicate 1 || | | G31 1-1 || Patient 1, replicate 1 || 96107 | ||
|- | |- | ||
| G31 1-2 || Patient 1, replicate 2 || | | G31 1-2 || Patient 1, replicate 2 || 96107 | ||
|- | |- | ||
| G31 1-3 || Patient 1, replicate 3 || | | G31 1-3 || Patient 1, replicate 3 || 96107 | ||
|- | |- | ||
| G31 2-1 || Patient 2, replicate 1 || | | G31 2-1 || Patient 2, replicate 1 || 74312 | ||
|- | |- | ||
| G31 2-2 || Patient 2, replicate 2 || | | G31 2-2 || Patient 2, replicate 2 || 74312 | ||
|- | |- | ||
| G31 2-3 || Patient 2, replicate 3 || | | G31 2-3 || Patient 2, replicate 3 || 74312 | ||
|} | |} | ||
Revision as of 02:29, 29 October 2014
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||||||||||||||||||||||||||||||
|
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
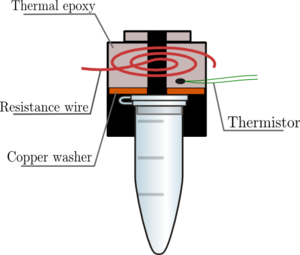
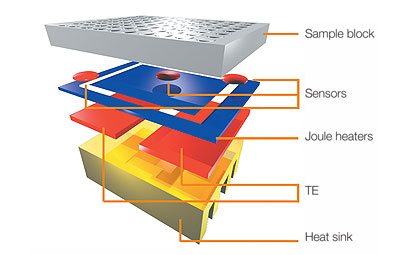
HEATED LID: 100°C INITIAL STEP: 95°C for 2 minutes NUMBER OF CYCLES: 35 Denature at 95°C for 30 seconds, Anneal at 57°C for 30 seconds, and Extend at 72°C for 30 seconds FINAL STEP: 72°C for 2 minutes FINAL HOLD: 4°C N/A. DNA Engine® Multi-Bay Thermal Cyclers — Alpha™ Unit Reaction Modules. Digital Image. Bio-Rad. Bio-Rad Laboratories Inc., N/A. Web. 10/27/14. The thermal cycling program multiplies a certain DNA sequence by an enormous amount. This occurs using a combination of primers, Taq Polymerase, and rapid heating and cooling. The program has two main settings: Primer Annealing and Primer Extension. The Annealing stage changes depending on sequence content, length of sequence, and primer concentration. The other setting, Extension, will change based on length of sequence and temperature. The amount of cycles depends on the amount of DNA to begin with.
Research and DevelopmentPCR - The Underlying Technology What is the function of each component of a PCR reaction? The template DNA in a PCR reaction contains the starting sequence of nucleotides that are to be amplified. The DNA sequence is added first and denatured by increasing temperature to about 95°C. There are now two separate strands from the original DNA double helix strand. Each strand will be used for replication to amplify the desired DNA sequence. Primers are small sequences of nucleotides that have been designed in a laboratory. Their function is to find the locus at which DNA replication is to occur, and bind to that location. Because primers are manufactured, they can be manipulated to have the exact desired sequence. To ensure that they don't attach to another area on the DNA strand, a length of about 20 nucleotide is used. A total of two primers are actually used in the reaction, one for each strand of the template DNA. These primers, after pairing with complimentary bases, kick-start the DNA polymerase replication, because DNA polymerase can only attach to certain start areas in the DNA. Taq Polymerase is an enzyme that will extend the length of the primers by adding complimentary nucleotides to the primer. This particular type of polymerase is used because of its heat tolerance, which can withstand well above 72°C, which is temperature that will allow extension but not annealing. Deoxyribonucleotides (dNTPs) are part of the basic units of DNA. There are four "bases," which are marked A for adenine, T for thymine, C for cytosine, and G for guanine. These four bases form sequences that translate as code and meaningful information in DNA. What happens to the components during each step of thermal cycling? Initial Step: 95°C for 3 minutes: Template DNA is denatured so that two strands result from the initial double helix. Denature at 95°C for 30 seconds: DNA denaturation is ensured so that the DNA strands will not reform. Primers will have been added by this point. Anneal at 57°C for 30 seconds: Two primers will bind will top and bottom strands of DNA, keeping the two strands separate while preparing for replication. Extend at 72°C for 30 seconds: Taq polymerase will have been added at this point, and will begin assembling the new DNA sequence from the two primers. At this temperature, no more annealing can occur, but DNA replication can, due to Taq's properties. Final Step at 72°C for 3 minutes: Full extension of the DNA sequence is ensured. Final Hold at 4°C: Newly formed sequences are allowed to solidify bonds and strengthen their hold on individual nucleotides. Base pairing A (adenine) pairs with T (thymine) C (cytosine) pairs with G (guanine) During which steps of thermal cycling does base-pairing occur? Annealing: primers attach to the two strands of DNA, pairing to a matching sequence in the template DNA. Extension: Taq polymerase brings over nucleotides, extending the primers and pairing them in order of the DNA sequence.
| ||||||||||||||||||||||||||||||||||