Biomolecular Breadboards: Difference between revisions
No edit summary |
|||
| Line 2: | Line 2: | ||
A ''biomolecular breadboard'' is a system that is designed to allow certain features of a circuit to be tested in a carefully controlled setting. These breadboards can be used to implement, debug, and characterize a wide variety of circuits, including both ''in vivo'' and ''in vitro'' devices. This page contains an overview of different biomolecular breadboards that are available. | A ''biomolecular breadboard'' is a system that is designed to allow certain features of a circuit to be tested in a carefully controlled setting. These breadboards can be used to implement, debug, and characterize a wide variety of circuits, including both ''in vivo'' and ''in vitro'' devices. This page contains an overview of different biomolecular breadboards that are available. | ||
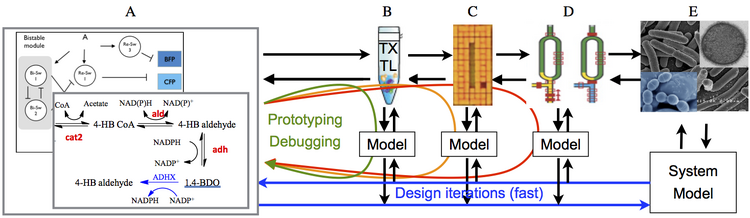
The figure below provides an overview of the basic breadboarding process. At the left is a circuit that we wish to implement and transform into a cell or other bimolecular chassis. Rather than try to directly get the circuit working in the cell, which requires time consuming iterations and difficult debugging, we instead use a sequence of simpler test environments ("breadboards"), where we can do much more rapid iterations between experiments, modeling and design. | The figure below provides an overview of the basic breadboarding process. At the left is a circuit that we wish to implement and transform into a cell or other bimolecular chassis. Rather than try to directly get the circuit working in the cell, which requires time consuming iterations and difficult debugging, we instead use a sequence of simpler test environments ("breadboards"), where we can do much more rapid iterations between experiments, modeling and design. | ||
| Line 23: | Line 21: | ||
=== DNA origami breadboard === | === DNA origami breadboard === | ||
A second breadboard family that we are developing explores the role of spatial localization and single molecule effects. For this, we are using | |||
DNA origami to spatially organize, study, and optimize existing ''in vitro'' transcription circuits such as oscillators. There are two kinds of spatial | |||
organization that can be effected by DNA origami. The first kind of organization, ''local patterning'' is intrinsic to the DNA origami themselves. For example, a rectangular origami provides a roughly 100~nm x 70~nm surface onto which other molecules can be positioned with a resolution of about 6 nanometers. | |||
A second kind of spatial organization afforded by DNA orgami is that of ''global placement'. We are developing a method for placing individual DNA origami at lithographically-patterned sites on a silicon dioxide surface: sticky patches in the shape of DNA origami are created by e-beam, origami are washed over the surface, and they bind and orient on the sticky patches. Because of the regular placement of origami, we expect to eventually be able to observe up to 1600 (40 x 40) origami localized circuits in parallel to achieve good statistics. | |||
=== Artificial cells === | === Artificial cells === | ||
=== Biochemical wires === | === Biochemical wires === | ||
Revision as of 04:03, 17 July 2012
| Home | Protocols | DNA parts | Preliminary Data | Models | More Info |
A biomolecular breadboard is a system that is designed to allow certain features of a circuit to be tested in a carefully controlled setting. These breadboards can be used to implement, debug, and characterize a wide variety of circuits, including both in vivo and in vitro devices. This page contains an overview of different biomolecular breadboards that are available.
The figure below provides an overview of the basic breadboarding process. At the left is a circuit that we wish to implement and transform into a cell or other bimolecular chassis. Rather than try to directly get the circuit working in the cell, which requires time consuming iterations and difficult debugging, we instead use a sequence of simpler test environments ("breadboards"), where we can do much more rapid iterations between experiments, modeling and design.
- DARPA Living Foundries kickoff presentation, 12 Jul 2012
Cell-free circuit breadboard
The cell-free circuit breadboard family is a sequence of in vitro protocols that can be used to test transcription and translation (TX-TL) circuits in a set of systematically-constructed environments that explore different elements of the external conditions in which the circuits must operate. This breadboard is based on the work of Vincent Noireaux at U. Minnesota. The transcription and translation machineries are extracted from E. coli cells (Shin and Noireaux, 2010). The endogenous DNA and mRNA from the cells is eliminated during the preparation. The resulting protein synthesis machinery is used to program cell-free TX-TL gene circuits in reactions of typical volume 10 $\mu$l. The gene circuits are engineered in the laboratory using standard molecular cloning techniques.
Breadboard features:
- The cell-free expression breadboard uses the E. coli housekeeping transcriptional machinery (σ70 and core RNA polymerase) to express all the other transcription factors, therefore providing all of the E. coli DNA regulatory components.
- The reaction buffer is entirely custom-made from pure chemicals. The concentration of energy molecules, ions, amino acids, molecular crowding agents, and other chemicals can be adjusted, as well as the pH of the reaction, over a wide range of concentrations.
- The breadboard can express proteins from either circular (plasmid) or linear DNA. PCR products can be used with some preparation.
- No antibiotic resistance markers are required, allowing plasmids using the same backbone to be included with different components. The concentrations of different plasmids can be modulated by changing the concentration of the plasmid.
- Approximate cost: $0.03/ul. Typical reactions are 10-20 ul ($0.30-0.60 per reaction)
DNA origami breadboard
A second breadboard family that we are developing explores the role of spatial localization and single molecule effects. For this, we are using DNA origami to spatially organize, study, and optimize existing in vitro transcription circuits such as oscillators. There are two kinds of spatial organization that can be effected by DNA origami. The first kind of organization, local patterning is intrinsic to the DNA origami themselves. For example, a rectangular origami provides a roughly 100~nm x 70~nm surface onto which other molecules can be positioned with a resolution of about 6 nanometers.
A second kind of spatial organization afforded by DNA orgami is that of global placement'. We are developing a method for placing individual DNA origami at lithographically-patterned sites on a silicon dioxide surface: sticky patches in the shape of DNA origami are created by e-beam, origami are washed over the surface, and they bind and orient on the sticky patches. Because of the regular placement of origami, we expect to eventually be able to observe up to 1600 (40 x 40) origami localized circuits in parallel to achieve good statistics.