Biomolecular Breadboards:Models
| Home | Protocols | DNA parts | Preliminary Data | Models | More Info |
This page contains information about models for simulation and analysis of biomolecular breadboard circuits. Additional information can be found in the following conference paper:
- An In Silico Modeling Toolbox for Rapid Prototyping of Circuits in a Biomolecular “Breadboard” System, Zoltan A. Tuza, Vipul Singhal, Jongmin Kim, Richard M. Murray. Submitted, 2013 Conference on Decision and Control (CDC).
TX-TL Toolbox for MATLAB
The TX-TL toolbox for MATLAB is a set of MATLAB functions that are designed to simplify the modeling of circuits used in the TX-TL cell free expression system. The source code for the toolbox is available via SourceForge.
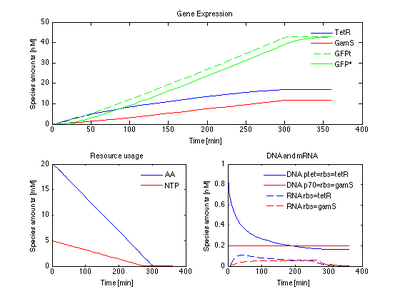
Simple example: Gene expression
This example shows constitutive expression of the destabilized enhanced Green Fluorescent Protein (deGFP) from a gene on a plasmid.
Create a tube containing TX-TL extract, 'E6'. The extract contains RNAP, ribosomes, sigma factors exonucleases and RNA-ase.
tube1 = txtl_extract('E6');
Create a tube containing TX-TL buffer, 'E6'. This contains amino acids and nucleotide pairs.
tube2 = txtl_buffer('E6');
Creat a new tube that will contain the DNA to be expressed, and name it 'circuit'.
tube3 = txtl_newtube('circuit');
Use txtl_add_dna to add custom deGFP DNA to tube3, under control of a constitutive p70 promoter:
dna_deGFP = txtl_add_dna(tube3, 'p70(50)', 'rbs(20)', 'deGFP(1000)', 4.0, 'plasmid');
The numbers in brackets are the lengths of the respective regions in number of base pairs (BP). These are used to calculate reaction rates for transcription, translation and degradation. The number 4.0 specifies that the initial concentration of the DNA (in nM) in the experiment once all the tubes have been combined. The last input argument species that the DNA is placed on a plasmid, and thus is not susceptible to exonuclease action.
Combine the tubes containing the extract, buffer and circuit into a final tube (referred to as Mobj) in a fixed ratio defined in the experimental protocol.
Mobj = txtl_combine([tube1, tube2, tube3]);
Define simulation time and set the 'StopTime' property of the simulation:
simulationTime = 15*60*60; configsetObj = getconfigset(Mobj, 'active'); set(configsetObj, 'StopTime', simulationTime);
Every model (tube) has an associated 'configuration set object' used to define how the model is to be simulated, and we can set its proterties as above. Simulate the model:
[simData] = txtl_runsim(Mobj,configsetObj); t_ode = simData.Time; x_ode = simData.Data;
This simulates the model object using the associated configuration set, and returns the time evolution of the concentration of the species in the model. Produce a standard plot:
txtl_plot(t_ode,x_ode,Mobj);
TXTL naming conventions
The TXTL toolbox uses a set of standard names for molecular species and compounds. If new components are added to the system, they must follow these naming conventions.
DNA, RNA and protein names
In order to allow automatic processing of reactions, the DNA, mRNA and proteins associated with a given gene have specific naming conventions:
- DNA: [DNA prom--utr--gene]
- mRNA: [RNA utr--gene]
- Protein: [protein gene]
The domain names for promoter (prom), untranslated region (utr), and gene can be arbitrary strings, but should generally consist of letters and numbers. DNA domains is specified as "dom1(len1)-dom2(len2)-..." where dom1 is a valid variable name and "(len1)" is an optional construct defining the length of the domain (in nucleotides). If length is not specified, then default value is used.
Standard species
A number of species are defined by the TXTL library and used to implement core processes:
- RNAP70 [nM]: core RNAP polymerase bound to sigma70
- Ribo [nM]: Ribosomes
- AA [unit]: amino acid "units". One unit of amino acid is equal to 100 nM. [AA] represents the total amino acid concentration.
- NTP [unit]: necleotide "units". One unit of nucleotide is equal to 100 nM. [NTP] represents the total concentration of nucleotides.
- RecBCD [nM]: exonuclease used to model degradation of linear DNA
Standard complexes
As part of the reactions that are set up by the toolbox, a number of standard species are created:
- [RNAP70:DNA prom--utr--gene] - RNA polymerase bound to DNA, corresponding roughly to the closed complex
- [NTP:RNAP70:DNA prom--utr--gene] - RNA polymerase bound to DNA, with NTP utilization (used for modeling transcription)
- [Ribo:RNA utr--gene] - Ribosome bound to mRNA
- [AA:Ribo:RNA utr--gene] - Ribosome bound to mRNA, with amino acid utilization (used for modeling translation)
- [RecBCD:DNA prom--utr--gene] - Exonuclease bound to DNA. This disables transcription.
TXTL toolbox functions
This section provides a summary of the main commands in the TXTL toolbox. More detailed documentation is available in the TXTL toolbox users guide (forthcoming) and via the MATLAB help function.
- txtl_extract - Create a tube containing the TXTL extract that is used for the reaction. The extract contains RNAP70, ribosomes and exonucleases. The extract batch is specified a string of the form 'eN' where N is an integer.
- txtl_buffer - Create a tube containing the buffer that is used for the reaction. The buffer contains NTPs and amino acids. The buffer batch is specified using a string of the form 'eN' where N is an integer.
- txtl_newtube - Create an empty tube in which DNA and other molecules can be places.
- txtl_dna - Put DNA into a tube. You must specify the promoter region, untranslated region (containing an RBS), and the gene (possibly with a degradation sequence)
- txtl_addspecies - Add a given species to a tube, with a given concentration.
- txtl_combine - Combine the contents of multiple tubes. Requires a list of tubes and a list of volumes (nominally in mL, but only relative concentrations matter)
TXTL components
Promoters
- p70 - default constitutive promoter
- ptet - promoter repressed by TetR
UTRs
- rbs - default ribosome binding site
Genes
- deGFP - GFP from the control plasmid. Creates both GFP and GFP* (mature)
- gamS - GamS protein, used to sequester RecBCD (exonuclease)
- tetR - TetR, including sequestration reaction with aTc
Additional Models
In addition to the TX-TL toolbox, several additional models have been developed for individual breadboard projects.
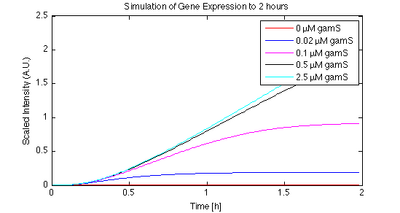
GamS modeling
For linear DNA, GamS can be used to reduce the rate of degradation of DNA and increase the expression level (see preliminary data for more information). The figure below shows the results of a preliminary model that we are developing to capture the affects of gamS.
Figure 1. Protein expression as a function of gamS concentration. The simulation results show the output of the model in 'protsynt81bis.m', generated with the script 'gamS_plot.m'. |