SBB12Ntbk-JonathanKotker: Difference between revisions
No edit summary |
|||
| Line 362: | Line 362: | ||
The procedure is described [http://openwetware.org/wiki/Template:SBB-Protocols_Micro3 here] and is used to extract and purify the DNA retrieved from the cells. This was used on the colonies that I picked [http://openwetware.org/wiki/SBB12Ntbk-JonathanKotker#Jon_Kotker.2C_1_March_2012 yesterday]. | The procedure is described [http://openwetware.org/wiki/Template:SBB-Protocols_Micro3 here] and is used to extract and purify the DNA retrieved from the cells. This was used on the colonies that I picked [http://openwetware.org/wiki/SBB12Ntbk-JonathanKotker#Jon_Kotker.2C_1_March_2012 yesterday]. | ||
==[[User:Jonathan_Kotker|Jon Kotker]], 6 March 2012== | |||
'''Part pBca9525-sbb1223''': ''Analytical Digests for Mapping'' | |||
The procedure is described [http://openwetware.org/wiki/Template:SBB-Protocols_Enz6 here] and is used to guess if the purified product is indeed the DNA sequence we need. We pick particular restriction enzymes such that the resulting fragments are of significantly different sizes, which should thus show up as two distinct bands on the analytical gel. If the analytical gel shows bands at the approximately proper locations, then we can send the purified product for digestion. | |||
Based on the construction file for this part, I chose the restriction enzymes EcoRI and BamHI, which should result in parts of length 1400bp and 2472bp. The procedure is modified accordingly: | |||
Recipe: | |||
* 4uL ddH2O | |||
* 4uL Miniprepped plasmid | |||
* 1uL 10x NEB Buffer 2 | |||
* 0.5uL EcoRI | |||
* 0.5uL BamHI | |||
Procedure: | |||
# Incubate at 37 degrees on the thermocycler for 30 minutes after 10:48am. (I took it out at 11:50am.) | |||
Revision as of 19:50, 6 March 2012
Jon Kotker, 13 February 2012
Hello World. Eat your vegetables.
Jon Kotker, 16 February 2012
First day of cloning and actual science. :)
Construction files here.
Part pBca9525-sbb1223: Preparing the mixture for PCA1
Ingredients given:
- Oligo mix at 100uM labeled "Caf mix 100"
- Forward oligo (caff.01.o01) at 10uM labeled "Caff 01.o01 10"
- Reverse oligo (caff.01.o14) at 10uM labeled "Caff 01.o14 10"
Recipe (for reference):
- 38uL ddH2O
- 5uL 10x expand buffer
- 5uL 2mM dNTPs
- 1uL oligo mixture "Caf mix 100" (100uM)
- 0.75uL Expand polymerase
Part pBca9525-sbb1204: Preparing the mixture for PCA1
Ingredients given:
- Oligo o1 at 100uM labeled "o1"
- Oligo o2 at 100uM labeled "o2"
- Oligo o4 at 100uM labeled "o4"
Recipe (for reference):
- 38uL ddH2O
- 5uL 10x expand buffer
- 5uL 2mM dNTPs
- 1uL oligo mixture (100uM)
- 0.75uL Expand polymerase
To obtain the oligo mixture, put (arbitrarily) 5uL each of "o1", "o2" and "o4" into an Eppendorf tube and mix.
Jon Kotker, 17 February 2012
Part pBca9525-sbb1223: Regular Zymo Cleanup of PCA assembly product
The procedure is described here but replicated below for completeness. It removes the polymerase, dNTPs, buffer, and most of the oligonucleotides, leaving behind the product of the first step of PCA (pca1 in the construction file).
- Add 180uL of Zymo ADB buffer to a 50uL reaction.
- Transfer into a Zymo column.
- Spin through for 15 seconds and discard waste.
- Add 200uL of Zymo Wash Buffer, which is basically 70% ethanol.
- Spin through for 15 seconds and discard waste.
- Add 200uL of Zymo Wash Buffer.
- Spin through for 15 seconds and discard waste.
- Spin through for 90 seconds at full speed to dry.
- Elute with 50uL of water (spinning for 60 seconds) into a fresh Eppendorf tube.
Note: After the third spin, I may have mixed up tubes with Greg Fukushima. There is a high chance that we got our correct tubes, but a significant chance that we did not. This is an important lesson in labeling my materials. Next time, I may attempt to restart this part, if the analytical gel reveals something potentially improper.
Part pBca9525-sbb1223: Amplification of PCA assembly product
The procedure is described here but replicated below for completeness. It starts the process of amplification of the correct full-length chunks (pca1 in the construction file).
Ingredients given:
- Forward oligo (caff.01.o01) at 10uM labeled "Caff 01.o01 10"
- Reverse oligo (caff.01.o14) at 10uM labeled "Caff 01.o14 10"
Recipe:
- 1uL each outer oligo at 10uM ("Caff 01.o01 10" and "Caff 01.o14 10")
- 1uL purified PCA product
- 0.5uL phusion
- 10uL 5x phusion buffer
- 5uL 2mM dNTPs
- 32.5uL ddH2O
Part pBca9525-sbb1204: Small-Fragment Zymo Cleanup of PCA assembly product
The procedure is described here but replicated below for completeness. It removes the polymerase, dNTPs, buffer, and most of the oligonucleotides, leaving behind the product of the first step of PCA (pca1 in the construction file).
- Add 100uL of Zymo ADB buffer to a 50uL reaction.
- Transfer into a Zymo column.
- Spin through for 15 seconds and discard waste.
- Add 200uL of Zymo Wash Buffer, which is basically 70% ethanol.
- Spin through for 15 seconds and discard waste.
- Add 200uL of Zymo Wash Buffer.
- Spin through for 15 seconds and discard waste.
- Spin through for 90 seconds at full speed to dry.
- Elute with 50uL of water (spinning for 60 seconds) into a fresh Eppendorf tube.
Note: Unfortunately, I did not have time to prepare the part pBca9525-sbb1204 for the second step of PCA, where the PCA assembly product gets amplified.
Jon Kotker, 21 February 2012
Part pBca9525-sbb1204: Amplification of PCA assembly product
The procedure is described here but replicated below for completeness. It starts the process of amplification of the correct full-length chunks (pca1 in the construction file).
Ingredients given:
- Forward oligo (o1) at 10uM labeled "o1 10"
- Reverse oligo (o2) at 10uM labeled "o2 10"
Recipe:
- 1uL each outer oligo at 10uM ("o1 10" and "o2 10")
- 1uL purified PCA product
- 0.5uL phusion
- 10uL 5x phusion buffer
- 5uL 2mM dNTPs
- 32.5uL ddH2O
Part pBca9525-sbb1223: Analytical Gel
An agarose gel electrophoresis procedure for analysis was run on 6uL of the PCA amplification product (with 2uL of blue dye), while a Zymo cleanup was performed on the rest. The procedure was used as a first pass to determine if the product is within the accepted range of the expected product; if not, the PCA gene synthesis should be restarted.
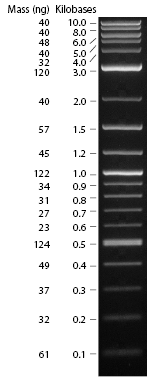
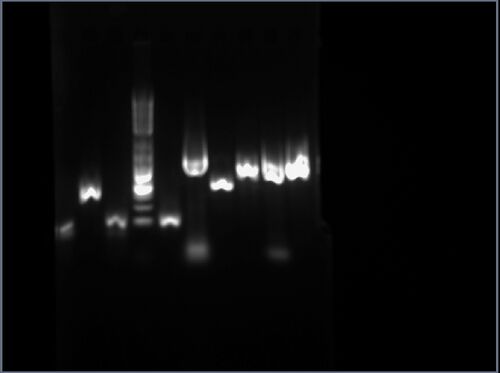
The result is shown here and replicated below for completeness:
My gel is the seventh lane from the left. Per Professor Anderson, there may have been some overloading of the dye, which could explain the runs on the side of the lanes. The ladder was placed in the fourth lane from the left. Based on this image, the product of the PCA amplification process seems to be around 500bp, when the expected product was 396bp long.
Part pBca9525-sbb1223: Regular Zymo Cleanup of PCA amplification product
The procedure is described here and replicated above.
Part pBca9525-sbb1223 (second): Preparing the mixture for PCA1
As described in the entry for February 17, there may have been a mix-up in parts. Just in case, I have prepared another mixture for PCA1 for part pBca9525-sbb1223. The procedure is detailed here.
Jon Kotker, 23 February 2012
Part pBca9525-sbb1223: Analytical Gel
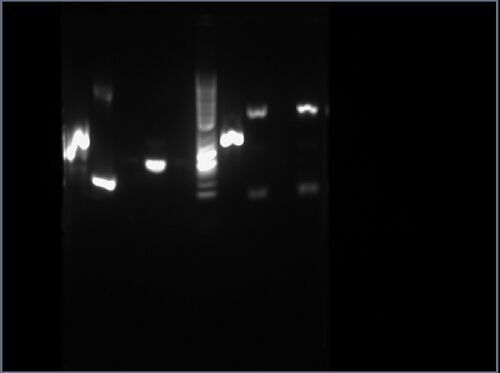
The result is shown here and replicated below for completeness:
My gel is the fourth lane from the left, having an approximate length between 500bp and 1000bp.
Part pBca9525-sbb1223: NheI/BamHI Digest of PCA amplification product
The procedure is described here, with NheI replacing EcoRI, but replicated below for completeness. It digests the amplification product (pca2 in the construction file), preparing it for ligation into the vector digest.
Recipe:
- 8uL of eluted PCR product
- 1uL of NEB Buffer 2
- 0.5uL of NheI
- 0.5uL of BamHI
Procedure:
- Incubate at 37 degrees on the thermocycler for one hour, starting at 10:13pm.
- Run an agarose gel and melt with 600uL ADB buffer at 55 degrees.
Part pBca9525-sbb1204: Analytical Gel
The result is shown here and replicated below for completeness:
My gel is the sixth lane from the left, having an approximate length between 100bp and 200bp, consistent with the expected length of 142bp.
Part pBca9525-sbb1204: Small-Fragment Zymo Cleanup of PCA amplification product
The procedure is described here and replicated above.
Part pBca9525-sbb1204: NheI/BamHI Digest of PCA amplification product
The procedure is described here, with NheI replacing EcoRI, but replicated below for completeness. It digests the amplification product (pca2 in the construction file), preparing it for ligation into the vector digest.
Recipe:
- 8uL of eluted PCR product
- 1uL of NEB Buffer 2
- 0.5uL of NheI
- 0.5uL of BamHI
Procedure:
- Incubate at 37 degrees on the thermocycler for one hour, starting at 11:09pm.
- Add 600uL ADB buffer.
Note: I misunderstood the directions and also added 250uL of isopropanol when I should not have, but Professor Anderson noted that it was all right and would not harm my product.
Part pBca9525-sbb1223 (second): Regular Zymo Cleanup of PCA assembly product
The procedure is described here and replicated above.
Part pBca9525-sbb1223 (second): Amplification of PCA assembly product
The procedure is described here and replicated above.
Jon Kotker, 24 February 2012

Part pBca9525-sbb1204: Small-Fragment Zymo Cleanup of digestion product
The procedure is described here and replicated above.
Part pBca9525-sbb1223: Regular Zymo Cleanup of digestion product
The procedure is described here and replicated above.
Part pBca9525-sbb1223 (second): Regular Zymo Cleanup of PCA amplification product
The procedure is described here and replicated above.
Jon Kotker, 28 February 2012
Part pBca9525-sbb1204: Ligation of NheI/BamHI Digests
The procedure is described here, but replicated below for completeness. It ligates the digested product of PCA amplification with a digested vector.
Recipe:
- 6.5uL ddH2O
- 1uL T4 DNA Ligase Buffer
- 1uL Vector digest
- 1uL Insert digest
- 0.5uL T4 DNA Ligase
Procedure:
- Pound upside down on the bench to mix.
- Give the mixture a quick spin to send it back to the bottom of the tube.
- Incubate on the benchtop for 30 minutes.
- Put on ice and proceed to the transformation.
Part pBca9525-sbb1204: Transformation by Heat-Shock
The procedure is described here, but replicated below for completeness.

- Thaw a 200uL aliquot of cells on ice.
- Add 50uL of water to the cells, if a greater volume is desired.
- Add 30uL of KCM to the cells.
- Put your ligation mixture on ice and let cool for a minute or two.
- Add 70uL of the cell cocktail to the ligation and stir to mix.
- Let sit on ice for 10 minutes.
- Heat shock for 90 seconds at 42 degrees (longer incubation may work better).
- Put back on ice for 1 minute.
- Add 100uL of 2YT and let shake in the 37 degree incubator for 1 hour.
- Add 70+uL on a plate with spectinomycin and let the plate incubate at 37 degrees overnight.
Part pBca9525-sbb1223: Ligation of NheI/BamHI Digests
The procedure is described here, but replicated below for completeness. It ligates the digested product of PCA amplification with a digested vector.
Recipe:
- 6.5uL ddH2O
- 1uL T4 DNA Ligase Buffer
- 1uL Vector digest
- 1uL Insert digest
- 0.5uL T4 DNA Ligase
Procedure:
- Pound upside down on the bench to mix.
- Give the mixture a quick spin to send it back to the bottom of the tube.
- Incubate on the benchtop for 30 minutes.
- Put on ice and proceed to the transformation.
Part pBca9525-sbb1223: Transformation by Heat-Shock
The procedure is described here, but replicated below for completeness.
- Thaw a 200uL aliquot of cells on ice.
- Add 50uL of water to the cells, if a greater volume is desired.
- Add 30uL of KCM to the cells.
- Put your ligation mixture on ice and let cool for a minute or two.
- Add 70uL of the cell cocktail to the ligation and stir to mix.
- Let sit on ice for 10 minutes.
- Heat shock for 90 seconds at 42 degrees (longer incubation may work better).
- Put back on ice for 1 minute.
- Add 100uL of 2YT and let shake in the 37 degree incubator for 1 hour.
- Add 70+uL on a plate with spectinomycin and let the plate incubate at 37 degrees overnight.
Jon Kotker, 1 March 2012
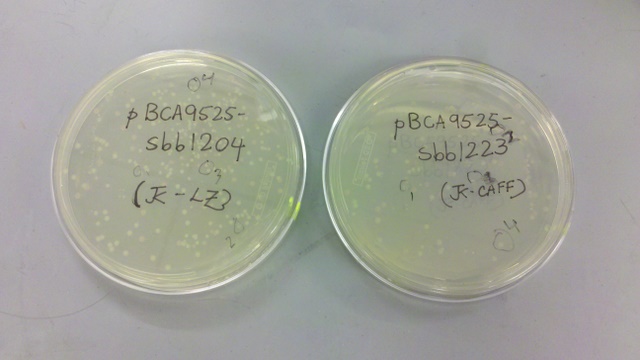
Part pBca9525-sbb1223: Picking of Colonies
The procedure is described here. However, I did not have to pick colonies: Zachary picked them for me and suspended them in media for growth. The colonies he chose are marked 1 and 2 on the plate. Nonetheless, he may have mislabeled the test tubes containing my cell mixture, but he mislabeled it as part "1233". Since there is no part "1233", those two test tubes are most likely mine.
Part pBca9525-sbb1223: Miniprep Purification of DNA
The procedure is described here and is used to extract and purify the DNA retrieved from the cells. To remind myself of the possible mislabeling, I labeled the corresponding Eppendorf tube with "1233" (instead of "1223"). Zachary suggested that I pick out more colonies for this part.
Part pBca9525-sbb1204: Picking of Colonies
The procedure is described here. However, I did not have to pick colonies: Zachary picked them for me and suspended them in media for growth. The colonies he chose are marked 1 and 2 on the plate.

Part pBca9525-sbb1204: Miniprep Purification of DNA
The procedure is described here and is used to extract and purify the DNA retrieved from the cells. It is replicated below for completeness.
- Pellet 2mL saturated culture by spinning full speed, 30 seconds in a 2mL Microcentrifuge tube.
- Dump supernatant.
- Add 250uL of P1 buffer into each tube. Resuspend the cells thoroughly using a vortexer.
- Add 250uL of P2 buffer, a base that denatures everything and causes cells to lyse. Gently mix up and down. The solution should become clearer and blue.
- Add 350uL of N3 buffer, an acid of pH ~5 that causes cell junk, including protein and chromosomal DNA, to precipitate, and leaves plasmids and other small molecules in solution. Slowly invert a few times, then shake.
- Spin in centrifuge at top speed for 5 minutes.
- Label blue columns with an alcohol-resistant lab pen.
- Pour liquid into columns and place the columns into the centrifuge. Spin at full speed for 15 seconds.
- Dump liquid out of the collectors under the columns. The DNA should be stuck to the white resin.
- Wash each column with 500uL of PB buffer.
- Spin in centrifuge at full speed for 15 seconds and flick out the liquid again.
- Wash with 750uL of PE buffer, which washes the salts off the resins.
- Spin in centrifuge at full speed for 15 seconds and flick out the liquid again.
- Spin in centrifuge at full speed for 90 sec to dry off all water and ethanol.
- Label new Microcentrifuge tubes and put columns in them.
- Elute them by adding 50uL of water down the middle of the column. Don't let it stick to the sides.
- Spin in centrifuge at top speed for 30 seconds.
- Take out the columns and cap the tubes.
Part pBca9525-sbb1223: Picking of Colonies
The procedure is described here and replicated below, albeit slightly modified in step 3, for completeness. I performed another picking in case the test tubes marked "1233", as noted earlier, were not mine.
- For each construct you will pick and later miniprep 2 colonies: I picked the colonies marked 3 and 4 on the plate by Zachary for overnight incubation.
- Add 4mL of LB media with the appropriate antibiotics to a clean test tube.
- Pick a well-isolated, round, and "normal" looking colony with a pipette tip.
- Drop it in the test tube.
- Incubate at 37 overnight.
Jon Kotker, 2 March 2012
Part pBca9525-sbb1204: Analytical Digests for Mapping
The procedure is described here and is used to guess if the purified product is indeed the DNA sequence we need. We pick particular restriction enzymes such that the resulting fragments are of significantly different sizes, which should thus show up as two distinct bands on the analytical gel. If the analytical gel shows bands at the approximately proper locations, then we can send the purified product for digestion.
Based on the construction file for this part, I chose the restriction enzymes EcoRI and XhoI, which should result in parts of length 2634bp and 986bp. The procedure is modified accordingly:
Recipe:
- 4uL ddH2O
- 4uL Miniprepped plasmid
- 1uL 10x NEB Buffer 2
- 0.5uL EcoRI
- 0.5uL XhoI
Procedure:
- Incubate at 37 degrees on the thermocycler for 30 minutes after 1:08pm.
Unfortunately, I only got as far as digesting some of the purified product, since there were no gels available.
Part pBca9525-sbb1223: Analytical Digests for Mapping
The procedure is described here and is used to guess if the purified product is indeed the DNA sequence we need. We pick particular restriction enzymes such that the resulting fragments are of significantly different sizes, which should thus show up as two distinct bands on the analytical gel. If the analytical gel shows bands at the approximately proper locations, then we can send the purified product for digestion.
Based on the construction file for this part, I chose the restriction enzymes EcoRI and BamHI, which should result in parts of length 1400bp and 2472bp. The procedure is modified accordingly:
Recipe:
- 4uL ddH2O
- 4uL Miniprepped plasmid
- 1uL 10x NEB Buffer 2
- 0.5uL EcoRI
- 0.5uL BamHI
Procedure:
- Incubate at 37 degrees on the thermocycler for 30 minutes after 1:08pm.
Unfortunately, I only got as far as digesting some of the purified product, since there were no gels available.
Part pBca9525-sbb1223: Miniprep Purification of DNA
The procedure is described here and is used to extract and purify the DNA retrieved from the cells. This was used on the colonies that I picked yesterday.
Jon Kotker, 6 March 2012
Part pBca9525-sbb1223: Analytical Digests for Mapping
The procedure is described here and is used to guess if the purified product is indeed the DNA sequence we need. We pick particular restriction enzymes such that the resulting fragments are of significantly different sizes, which should thus show up as two distinct bands on the analytical gel. If the analytical gel shows bands at the approximately proper locations, then we can send the purified product for digestion.
Based on the construction file for this part, I chose the restriction enzymes EcoRI and BamHI, which should result in parts of length 1400bp and 2472bp. The procedure is modified accordingly:
Recipe:
- 4uL ddH2O
- 4uL Miniprepped plasmid
- 1uL 10x NEB Buffer 2
- 0.5uL EcoRI
- 0.5uL BamHI
Procedure:
- Incubate at 37 degrees on the thermocycler for 30 minutes after 10:48am. (I took it out at 11:50am.)