Beauchamp:Retinotopy: Difference between revisions
No edit summary |
No edit summary |
||
| Line 23: | Line 23: | ||
Here are some processing steps to analyse this data in AFNI: | Here are some processing steps to analyse this data in AFNI: | ||
First step: measure motion in each run separately, perform the registration, save motion parameters | |||
create average EPI image to register to | |||
3dTcat -prefix {$ec}rall {$ec}r1+orig.HEAD {$ec}r2+orig.HEAD {$ec}r3+orig.HEAD {$ec}r4+orig.HEAD {$ec}r5+orig.HEAD \ | 3dTcat -prefix {$ec}rall {$ec}r1+orig.HEAD {$ec}r2+orig.HEAD {$ec}r3+orig.HEAD {$ec}r4+orig.HEAD {$ec}r5+orig.HEAD \ | ||
{$ec}r6+orig.HEAD {$ec}r7+orig.HEAD {$ec}r8+orig.HEAD | {$ec}r6+orig.HEAD {$ec}r7+orig.HEAD {$ec}r8+orig.HEAD | ||
| Line 51: | Line 51: | ||
remove quadratic trend from each run separately | |||
foreach r (1 2 3 4 5 6 7 8) | foreach r (1 2 3 4 5 6 7 8) | ||
3dDetrend -prefix {$ec}r{$r}vrdt -polort 2 {$ec}r{$r}vr+orig | 3dDetrend -prefix {$ec}r{$r}vrdt -polort 2 {$ec}r{$r}vr+orig | ||
end | end | ||
blur slightly | |||
foreach r (1 2 3 4 5 6 7 8) | foreach r (1 2 3 4 5 6 7 8) | ||
3dmerge -doall -1blur_rms 3 -prefix {$ec}r{$r}vrdtbl {$ec}r{$r}vrdt+orig | 3dmerge -doall -1blur_rms 3 -prefix {$ec}r{$r}vrdtbl {$ec}r{$r}vrdt+orig | ||
| Line 61: | Line 61: | ||
rm junk* | rm junk* | ||
foreach r (1 2 3 4 5 6 7 8) | foreach r (1 2 3 4 5 6 7 8) | ||
Convert to float as required by DFT | |||
3dcalc -prefix junkr{$r} -datum float -a {$ec}r{$r}vrdtbl+orig -expr "a" | 3dcalc -prefix junkr{$r} -datum float -a {$ec}r{$r}vrdtbl+orig -expr "a" | ||
perform FFT on each run separately | |||
3dDFT -prefix {$ec}r{$r}vrdtblFFT -detrend junkr{$r}+orig | 3dDFT -prefix {$ec}r{$r}vrdtblFFT -detrend junkr{$r}+orig | ||
end | end | ||
| Line 75: | Line 75: | ||
end | end | ||
Average magnitude squared across runs | |||
3dMean -prefix {$ec}rallvrdtblFFTMAG {$ec}r?vrdtblFFTMAG+orig.HEAD | 3dMean -prefix {$ec}rallvrdtblFFTMAG {$ec}r?vrdtblFFTMAG+orig.HEAD | ||
Calculate Fratio between frequency of interest and neighboring frequencies | |||
rm junk* | rm junk* | ||
3dcalc -prefix junkMAGSTAT -a3 {$ec}rallvrdtblFFTMAG+orig -b4 {$ec}rallvrdtblFFTMAG+orig -c5 {$ec}rallvrdtblFFTMAG+orig \ | 3dcalc -prefix junkMAGSTAT -a3 {$ec}rallvrdtblFFTMAG+orig -b4 {$ec}rallvrdtblFFTMAG+orig -c5 {$ec}rallvrdtblFFTMAG+orig \ | ||
-d6 {$ec}rallvrdtblFFTMAG+orig -e7 {$ec}rallvrdtblFFTMAG+orig -expr "c^2 / ( (a^2 + b^2 + d^2 + e^2)/4 )" | -d6 {$ec}rallvrdtblFFTMAG+orig -e7 {$ec}rallvrdtblFFTMAG+orig -expr "c^2 / ( (a^2 + b^2 + d^2 + e^2)/4 )" | ||
Convert to fbuc | |||
3dbucket -prefix {$ec}rallvrdtblFFTMAGSTAT -fbuc junkMAGSTAT+orig | 3dbucket -prefix {$ec}rallvrdtblFFTMAGSTAT -fbuc junkMAGSTAT+orig | ||
Give it correct stats--numerator degrees of freedom is 2*number of runs, denominator degrees of freedom is 4*number of runs | |||
3drefit -sublabel 0 Significance -substatpar 0 fift 16 64 -subrepkey 0 Significance {$ec}rallvrdtblFFTMAGSTAT+orig | 3drefit -sublabel 0 Significance -substatpar 0 fift 16 64 -subrepkey 0 Significance {$ec}rallvrdtblFFTMAGSTAT+orig | ||
Calculate appropriate phases | |||
flip CW and CON runs | |||
foreach r (3 4 7 8) | foreach r (3 4 7 8) | ||
3dcalc -datum short -prefix {$ec}r{$r}vrdtblFFTf5flip -a {$ec}r{$r}vrdtblFFTf5+orig -expr "-a" | 3dcalc -datum short -prefix {$ec}r{$r}vrdtblFFTf5flip -a {$ec}r{$r}vrdtblFFTf5+orig -expr "-a" | ||
3drefit -sublabel 0 FFTr{$r}f5flip -subrepkey 0 FFTr{$r}f5flip {$ec}r{$r}vrdtblFFTf5flip+orig | 3drefit -sublabel 0 FFTr{$r}f5flip -subrepkey 0 FFTr{$r}f5flip {$ec}r{$r}vrdtblFFTf5flip+orig | ||
end | end | ||
Average together | |||
rm junk* | rm junk* | ||
3dcalc -prefix junkeccavg -datum short -a {$ec}r2vrdtblFFTf5+orig -b {$ec}r4vrdtblFFTf5flip+orig \ | 3dcalc -prefix junkeccavg -datum short -a {$ec}r2vrdtblFFTf5+orig -b {$ec}r4vrdtblFFTf5flip+orig \ | ||
| Line 101: | Line 101: | ||
-c {$ec}r5vrdtblFFTf5+orig -d {$ec}r7vrdtblFFTf5flip+orig -expr "mean(a,b,c,d)" | -c {$ec}r5vrdtblFFTf5+orig -d {$ec}r7vrdtblFFTf5flip+orig -expr "mean(a,b,c,d)" | ||
Combine stats, individual runs, and averages in one file for ease of viewing | |||
3dbucket -prefix {$ec}Ret -fbuc {$ec}rallvrdtblFFTMAGSTAT+orig junkeccavg+orig junkpolavg+orig \ | 3dbucket -prefix {$ec}Ret -fbuc {$ec}rallvrdtblFFTMAGSTAT+orig junkeccavg+orig junkpolavg+orig \ | ||
{$ec}r{$r}vrdtblFFTf5+orig.HEAD {$ec}r?vrdtblFFTf5flip+orig.HEAD | {$ec}r{$r}vrdtblFFTf5+orig.HEAD {$ec}r?vrdtblFFTf5flip+orig.HEAD | ||
Revision as of 17:29, 30 November 2007
Beauchamp Lab wiki
|
Phase-Mapped Retinotopy
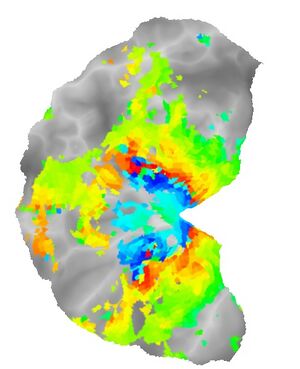
Visual areas are retinotopic. An efficient way to map visual areas is to present stimuli that traverse the visual field.
Here are three snapshots of an eccentricity mapping stimulus:



Over the course of a 5-minute scan series, the ring expands from central to peripheral five times.
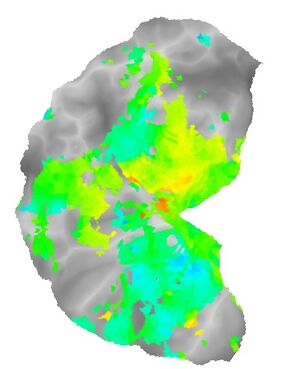
Here are three snapshots of a polar angle mapping stimulus:
Over the course of a 5-minute scan series, the wedge rotates around the fixation point five times.
Processing Phase-Mapped Retinotopy Data in AFNI
Here are some processing steps to analyse this data in AFNI:
First step: measure motion in each run separately, perform the registration, save motion parameters create average EPI image to register to
3dTcat -prefix {$ec}rall {$ec}r1+orig.HEAD {$ec}r2+orig.HEAD {$ec}r3+orig.HEAD {$ec}r4+orig.HEAD {$ec}r5+orig.HEAD \
{$ec}r6+orig.HEAD {$ec}r7+orig.HEAD {$ec}r8+orig.HEAD
3dTstat -mean -prefix {$ec}EPImean {$ec}rall+orig.HEAD
Motion Correction
for more info, see Motion and Distortion Correction
Alternative 1: Perform registration to mean image
foreach r (1 2 3 4 5 6 7 8)
3dvolreg -prefix {$ec}r{$r}vr -base {$ec}EPImean+orig -input {$ec}r{$r}+orig -1Dfile {$ec}r{$r}vr.1D -cubic
end
Alternative 2: For even greater precision, we Allineate to the hi-resolution anatomical image.
3dAllineate -base {$ec}anatSScrop+orig -1Dmatrix_save {$ec}EPI_to_T1.aff12.1D -input {$ec}EPImean+orig -EPI -cost mi
foreach r (1 2 3 4 5 6 7 8)
3dvolreg -prefix NULL -1Dmatrix_save {$ec}r{$r}EPI_to_EPI.aff12.1D -base {$ec}EPImean+orig -input {$ec}r{$r}+orig -cubic
cat_matvec {$ec}EPI_to_T1.aff12.1D {$ec}r{$r}EPI_to_EPI.aff12.1D > {$ec}r{$r}EPI_to_EPI_to_T1.aff12.1D
3dAllineate -master {$ec}anatSScrop+orig -mast_dxyz 2.0 -1Dmatrix_apply {$ec}r{$r}EPI_to_EPI_to_T1.aff12.1D \
-input {$ec}r{$r}+orig -final quintic -prefix {$ec}r{$r}vr+orig
end
remove quadratic trend from each run separately
foreach r (1 2 3 4 5 6 7 8)
3dDetrend -prefix {$ec}r{$r}vrdt -polort 2 {$ec}r{$r}vr+orig
end
blur slightly
foreach r (1 2 3 4 5 6 7 8)
3dmerge -doall -1blur_rms 3 -prefix {$ec}r{$r}vrdtbl {$ec}r{$r}vrdt+orig
end
rm junk*
foreach r (1 2 3 4 5 6 7 8)
Convert to float as required by DFT
3dcalc -prefix junkr{$r} -datum float -a {$ec}r{$r}vrdtbl+orig -expr "a"
perform FFT on each run separately
3dDFT -prefix {$ec}r{$r}vrdtblFFT -detrend junkr{$r}+orig
end
foreach r (1 2 3 4 5 6 7 8)
# Retinotopic stimulus has periodicity at frequency 5, calculate phase at only that frequency
3dcalc -datum short -prefix {$ec}r{$r}vrdtblFFTf5 -cx2r PHASE -a {$ec}r{$r}vrdtblFFT+orig'[5]' -expr "a"
3drefit -sublabel 0 FFTr{$r}f5 -subrepkey 0 FFTr{$r}f5 {$ec}r{$r}vrdtblFFTf5+orig
# Calculate magnitude squared at all frequencies
3dcalc -datum short -prefix {$ec}r{$r}vrdtblFFTMAG -cx2r ABS -a {$ec}r{$r}vrdtblFFT+orig -expr "a*a"
end
Average magnitude squared across runs
3dMean -prefix {$ec}rallvrdtblFFTMAG {$ec}r?vrdtblFFTMAG+orig.HEAD
Calculate Fratio between frequency of interest and neighboring frequencies
rm junk*
3dcalc -prefix junkMAGSTAT -a3 {$ec}rallvrdtblFFTMAG+orig -b4 {$ec}rallvrdtblFFTMAG+orig -c5 {$ec}rallvrdtblFFTMAG+orig \
-d6 {$ec}rallvrdtblFFTMAG+orig -e7 {$ec}rallvrdtblFFTMAG+orig -expr "c^2 / ( (a^2 + b^2 + d^2 + e^2)/4 )"
Convert to fbuc
3dbucket -prefix {$ec}rallvrdtblFFTMAGSTAT -fbuc junkMAGSTAT+orig
Give it correct stats--numerator degrees of freedom is 2*number of runs, denominator degrees of freedom is 4*number of runs
3drefit -sublabel 0 Significance -substatpar 0 fift 16 64 -subrepkey 0 Significance {$ec}rallvrdtblFFTMAGSTAT+orig
Calculate appropriate phases flip CW and CON runs
foreach r (3 4 7 8)
3dcalc -datum short -prefix {$ec}r{$r}vrdtblFFTf5flip -a {$ec}r{$r}vrdtblFFTf5+orig -expr "-a"
3drefit -sublabel 0 FFTr{$r}f5flip -subrepkey 0 FFTr{$r}f5flip {$ec}r{$r}vrdtblFFTf5flip+orig
end
Average together
rm junk*
3dcalc -prefix junkeccavg -datum short -a {$ec}r2vrdtblFFTf5+orig -b {$ec}r4vrdtblFFTf5flip+orig \
-c {$ec}r6vrdtblFFTf5+orig -d {$ec}r8vrdtblFFTf5flip+orig -expr "mean(a,b,c,d)"
3dcalc -prefix junkpolavg -datum short -a {$ec}r1vrdtblFFTf5+orig -b {$ec}r3vrdtblFFTf5flip+orig \
-c {$ec}r5vrdtblFFTf5+orig -d {$ec}r7vrdtblFFTf5flip+orig -expr "mean(a,b,c,d)"
Combine stats, individual runs, and averages in one file for ease of viewing
3dbucket -prefix {$ec}Ret -fbuc {$ec}rallvrdtblFFTMAGSTAT+orig junkeccavg+orig junkpolavg+orig \
{$ec}r{$r}vrdtblFFTf5+orig.HEAD {$ec}r?vrdtblFFTf5flip+orig.HEAD
Here are flat maps showing the thresholded eccentricity and polar angle maps produced by this analysis: