IGEM:Caltech/2007/Project/Riboregulator: Difference between revisions
From OpenWetWare
Jump to navigationJump to search
No edit summary |
No edit summary |
||
| Line 13: | Line 13: | ||
==RNA Background== | ==RNA Background== | ||
RNA has traditionally been considered a carrier of information between the gene and the final protein product. However, recent evidence is accumulating for alternate functions for RNA - namely, the use of RNA molecules as regulatory elements in the cell. Examples include ribozymes, siRNA, and riboswitches. | RNA has traditionally been considered a carrier of information between the gene and the final protein product. However, recent evidence is accumulating for alternate functions for RNA - namely, the use of RNA molecules as regulatory elements in the cell. Examples include ribozymes, siRNA, and riboswitches. | ||
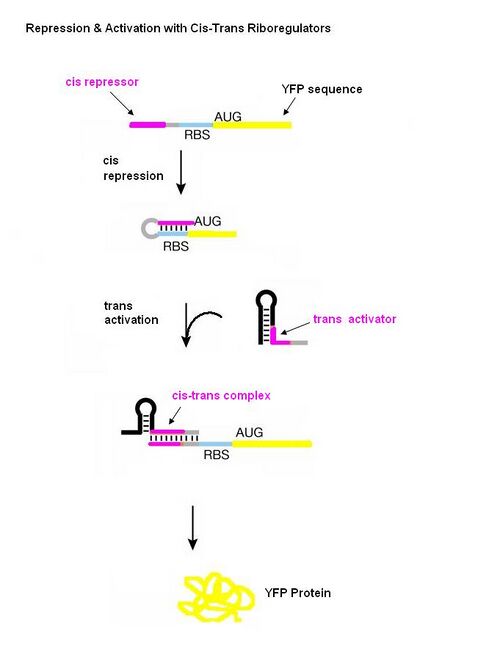
[[Image:Cis trans diagram.JPG|left|thumb|500px|Figure adopted from: Isaacs FJ, Dwyer DJ, Ding C, Pervouchine DD, Cantor CR, and Collins JJ. Engineered riboregulators enable post-transcriptional control of gene expression. Nat Biotechnol 2004 Jul; 22(7) 841-7.]] | |||
==Riboregulators== | ==Riboregulators== | ||
New insights into this regulatory role of RNA have allowed for engineering new RNA molecules which control cell behavior interactions with other RNAs or with small molecule ligands. One such engineering approach which enables post-transcriptional control of gene expression is the riboregulator developed by Collins and coworkers. The riboregulator is composed of two interacting parts, a cis-repressive sequence and a trans-activating sequence. The cis sequence is placed directly upstream of the ribosome binding site (RBS) of the gene regulated by the riboregulator and is complimentary to the RBS. This sequence forms a stem-loop structure in the 5’-untranslated region of the mRNA which prevents ribosome binding and thus translation. The trans RNA is complementary to the cis sequence. When the trans RNA is present, it binds to the cis sequence, the RBS site becomes exposed, and the ribosome can bind and translate the mRNA. | New insights into this regulatory role of RNA have allowed for engineering new RNA molecules which control cell behavior interactions with other RNAs or with small molecule ligands. One such engineering approach which enables post-transcriptional control of gene expression is the riboregulator developed by Collins and coworkers. The riboregulator is composed of two interacting parts, a cis-repressive sequence and a trans-activating sequence. The cis sequence is placed directly upstream of the ribosome binding site (RBS) of the gene regulated by the riboregulator and is complimentary to the RBS. This sequence forms a stem-loop structure in the 5’-untranslated region of the mRNA which prevents ribosome binding and thus translation. The trans RNA is complementary to the cis sequence. When the trans RNA is present, it binds to the cis sequence, the RBS site becomes exposed, and the ribosome can bind and translate the mRNA. | ||
| Line 29: | Line 33: | ||
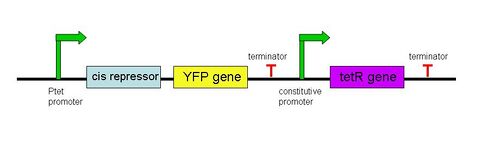
==Riboregulator Integration in Our Phage/''E. coli'' System== | ==Riboregulator Integration in Our Phage/''E. coli'' System== | ||
The riboregulator determines what happens to the cell once it is infected by phage. We aimed to design three types of riboregulated systems (one for each protein N, Q, and Cro). For instance, for the N protein-regulated system, the phage genome would have a mutant version of the N gene as well as an N gene suppressed with a cis-repressive sequence would be additionally integrated into the genome. When the phage infects a wild-type E.coli cell, nothing should happen. However, when it infects a cell with the trans-activating noncoding RNA, the cell should lyse because the RBS of the N gene is no longer sequestered by the cis-repressed RNA loop. | The riboregulator determines what happens to the cell once it is infected by phage. We aimed to design three types of riboregulated systems (one for each protein N, Q, and Cro). For instance, for the N protein-regulated system, the phage genome would have a mutant version of the N gene as well as an N gene suppressed with a cis-repressive sequence would be additionally integrated into the genome. When the phage infects a wild-type E.coli cell, nothing should happen. However, when it infects a cell with the trans-activating noncoding RNA, the cell should lyse because the RBS of the N gene is no longer sequestered by the cis-repressed RNA loop. | ||
| Line 57: | Line 58: | ||
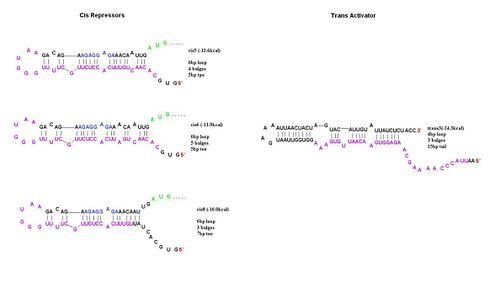
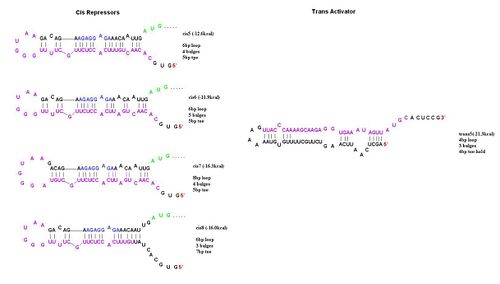
In order to determine the optimal level of repression, various aspects of RNA secondary structure were considered, including inner loops, single base pair bulges, and varying loop sizes. Higher free energies (i.e. less complementarity due to base pair mismatches) favor activation by taRNA because they destabilize the stem and facilitate the open RBS form upon addition of taRNA. The cis elements were inserted downstream of the Ptet promoter. The stem consisted of approximately 20-nt and the loop ranged from 6 to 10-nt. The YFP gene was inserted directly downstream of the cis sequence. Flow cytometry measurements were taken to quantify the expression of the cis riboregulated YFP gene. | In order to determine the optimal level of repression, various aspects of RNA secondary structure were considered, including inner loops, single base pair bulges, and varying loop sizes. Higher free energies (i.e. less complementarity due to base pair mismatches) favor activation by taRNA because they destabilize the stem and facilitate the open RBS form upon addition of taRNA. The cis elements were inserted downstream of the Ptet promoter. The stem consisted of approximately 20-nt and the loop ranged from 6 to 10-nt. The YFP gene was inserted directly downstream of the cis sequence. Flow cytometry measurements were taken to quantify the expression of the cis riboregulated YFP gene. | ||
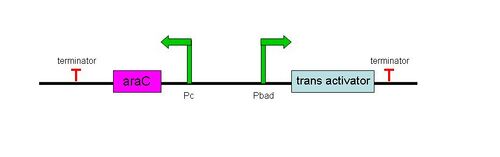
[[Image:Wiki overview2.JPG|right|thumb|500px|The general structure of a trans-activating element in our inducible Pbad/araC system.]] | |||
'''Trans Activation''' | '''Trans Activation''' | ||
To initiate translation, five trans-activating elements (ta1-ta5) were designed. Each sequence binds to the cis-repressive elements and opens up the RBS. For ta1-ta4, part of the sequence was complementary to the hairpin loop of the cr element and extending in the 5’ direction. Element ta5 was designed to bind to the 5’ end of the cis regulator and open up the hairpin in the 3’ direction. ta1 and ta2 were complimentary to cr1-4; ta3-5 were complimentary to cr5-8. The activation with these ta elements is currently being determined. | To initiate translation, five trans-activating elements (ta1-ta5) were designed. Each sequence binds to the cis-repressive elements and opens up the RBS. For ta1-ta4, part of the sequence was complementary to the hairpin loop of the cr element and extending in the 5’ direction. Element ta5 was designed to bind to the 5’ end of the cis regulator and open up the hairpin in the 3’ direction. ta1 and ta2 were complimentary to cr1-4; ta3-5 were complimentary to cr5-8. The activation with these ta elements is currently being determined. | ||
| Line 72: | Line 84: | ||
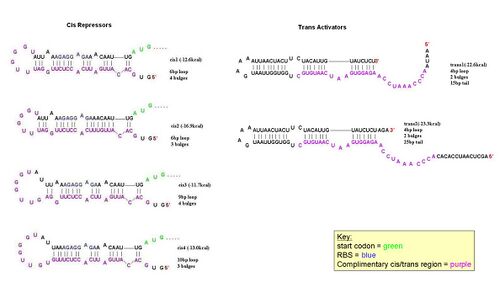
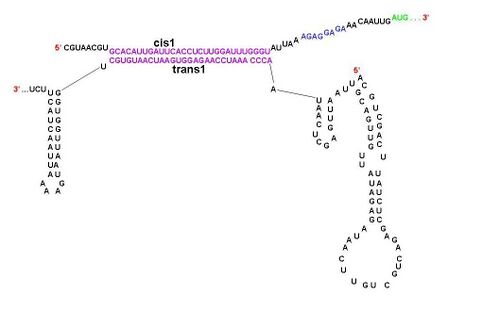
[[Image:Cis trans compliments.JPG|left|thumb|500px|A schematic diagram showing the parts of cis1 repressor and trans1 activator that are complimentary to each other. Note that the RBS is now open.]] | [[Image:Cis trans compliments.JPG|left|thumb|500px|A schematic diagram showing the parts of cis1 repressor and trans1 activator that are complimentary to each other. Note that the RBS is now open.]] | ||
==Experiments with Riboregulators== | ==Experiments with Riboregulators== | ||