Sarah Carratt: Week 6
Instructions
- List the state variables needed to model the process of interest.
- Propose at least one system of differential equations you think will model the dynamics.
- Discuss the terms in your equation(s) in order to justify your choices.
- List all parameters your model requires for numerical simulation.
- Discuss the relationship between the data in the papers by ter Schure et al and the state variables (and parameters).
Online Sources
Student Response
Variables Needed for a Model
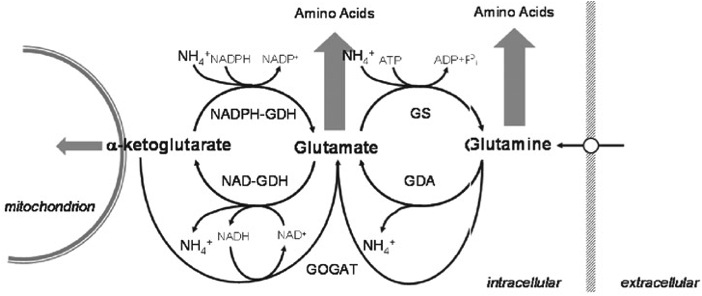
- ammonium → nitrogen
- α-ketogluterate
- glutamate
- glutamine
These four variables are the things that we will need to watch/model as they change over time. In the image, these variables can be seen in context of nitrogen metabolism.
Differential Equations and Discussion of Terms
[] = concentration of enclosed
D = dilution rate
u = feed concentration
k1, k2, k3, k4 = rate constants
Vmax = enzyme concentrations (constant)
L1, L2, L3, L4 = loss of state variable to outside factors/processes in cell and also because of the backwards conversions/cycle
- d[glutamine]/dt = D*u - Vmax([glutamine]/k1[glutamine])+ Vmax([glutamate]/k2[glutamate])- L1
- d[glutamate]/dt = D*u -Vmax([α-ketogluterate]/k3[α-ketogluterate]) + Vmax([α-ketogluterate]/k4[α-ketogluterate])- Vmax([glutamate]/k2[glutamate])+ Vmax([glutamine]/k1[glutamine])- L2
- d[α-ketogluterate]/dt = D*u-Vmax([α-ketogluterate]/k4[α-ketogluterate]) + Vmax([gluterate]/k3[gluterate]) - L3
- d[nitrogen]/dt = D*u + [ammonium] - L4
Parameters for Model
- Vmax (k*[enzymes]0: GDA, GS, NAD-GDH, NADPH-GDH)
- D (dilution rate) CONSTANT
- u (includes glucose/ammonium aka carbon/nitrogen)
- ammonium changes
- glucose is constant
Relationship between ter Schure et al and Parameters
All variables are connected to ter Schure. Originally, I was confused with how to include carbon/glucose, but I believe that it is accounted for in the feed concentration and dilution. I shouldn't need a fifth equation for glucose. The major difference between my parameters and ter Schure is that I have not focused on individual enzymes. I tried to factor them into my equation but I'm not sure they can be accounted for in the same ways.
Correct Answers
STATE VARIABLES:
- α-ketogluterate
- Glutamate
- Glutamine
- Ammonium → Nitrogen
WHAT IS THE SYSTEM?
- Cell
- Chemostat Reactor
UNITS:
- moles/volume
- moles/(volume*time)
EQUATIONS:
- d[α-ketogluterate]/dt = -V4([α-ketogluterate]/k4+[α-ketogluterate]) + V3([glutamate]/k3+[glutamate])
- d[glutamine]/dt = -V1([glutamine]/k1+[glutamine]) + V2([glutamate]/k2+[glutamate])
- d[glutamate]/dt = V1([glutamine]/k1+[glutamine])- V2([glutamate][ammonium]/k2+[glutamate][ammonium]) + V3([α-ketogluterate][ammonium]/k3+[α-ketogluterate][ammonium]) - V4([glutamate]/k4+[glutamate]) + V5([α-ketogluterate][glutamine]/k5+[α-ketogluterate][glutamine])
- d[ammonium]/dt = D*u + Va1([glutamine]/ka1+[glutamine])+ Va4([glutamate]/ka4+[glutamate])
EQUATIONS WITH SIMPLE VARIABLES:
- d[A]/dt = -V4([A]/k4+[A]) + V3([B]/k3+[B])
- d[B]/dt = V1([C]/k1+[C])- V2([B][D]/k2+[B][D]) + V3([A][D]/k3+[A][D]) - V4([B]/k4+[B]) + V5([A][C]/k5+[A][C])
- d[C]/dt = -V1([C]/k1+[C]) + V2([B]/k2+[B])
- d[D]/dt = D*u + Va1([C]/ka1+[C])+ Va4([B]/ka4+[B])
A=first substrate (α-ketogluterate), B=second substrate (glutamate), C=third substrate (glutamine), D=fourth substrate (ammonium)
NOTES:
- D*u = source, inflow (dilution rate*feed concentration)
- V# = enzyme level, accounts for loss, "hides amount of enzyme" (k*e0: GDA, GS, NAD-GDH, NADPH-GDH)
- the "L" constant is troubling in terms of units
- strategy: fit to orignial equations, E+S↔ES→E+P and E+P↔EP→E+S
- α-ketogluterate has no nitrogen, glutamate has one, glutamine has two
- food for thought: conserved? 2 substrate model="right"? what if you set d/dt=0 to look at equilibrium? use steady state to find constants?
Navigation Guide
Individual Assignments
Class Assignments
Class Notes
Internal Links
| BIOL398-01/S11:Assignments | BIOL398-01/S11:People | BIOL398-01/S11:Sarah Carratt |