SBOL Semantic
Semantic SBOL
Goals
- Implement the core data model described in The BioBricks Foundation:Standards/Technical/Exchange/Core Data Model as a simple OWL ontology.
- Provide access to data described using Semantic SBOL
Summary of Current State of Development
Updated: **Michal Galdzicki 03:23, 21 April 2010 (EDT)::
- Ontology: http://sbol.bhi.washington.edu/rdf/sbol.owl
- Data: http://sbol.bhi.washington.edu/data/SemanticSBOLv0.13_BioBrick_Data_v0.14.rdf (21M) (From Cesar/ Emma)
- Data: http://sbol.bhi.washington.edu/data/SemanticSBOLv0.13_BioBrick_Data_v0.15.rdf (partial data from Registry XML)
- Access Point: Sesame 1.27 http://sbol.bhi.washington.edu/sesame/ (Publicly Read Only)
- API prototype: Python sbol3.py
- Code: http://synbiolib.hg.sourceforge.net/hgweb/synbiolib/synbiolib/
- Documentation: http://sbol.bhi.washington.edu/py/html/
- Usage: To implement is to use!
Explanation
The initial SBOL effort is aimed towards providing a standard method of transferring the minimal information needed for the description of standardized parts for Synthetic Biology. To accomplish this goal in terms of data transfer over the network SBOL Semantic provides an implementation of the data model using W3C Semantic Web standards. The data model was encoded as an OWL-DL ontology. This simple ontology allows for the description of data elements in terms of Classes (i.e. Parts, Sequence Features, etc), the object properties, relationships, between them (ie. [has]feature, [has]annotation, etc) and data properties (i.e. name, author, short description, etc). The OWL definitions provide both structure and definition for the data. Data encoded using OWL is provided in RDF/XML syntax which allows the use of standard parsers and query languages to access the encoded information. RDF/XML syntax maybe parsed directly from the file form, or can be accessed using a query protocol (i.e. SPARQL).
Ontology Definition
The current version is both conveniently at the url equal to the namespace set in the ontology
Data
Data: http://sbol.bhi.washington.edu/data/SemanticSBOLv0.13_BioBrick_Data_v0.13.rdf (21M)(From Cesar/ Emma)
Access Point
Access Point: Sesame 1.27 http://sbol.bhi.washington.edu/sesame/ (Publicly Read Only)
API
API prototype: Written as python objects and methods for working with Semantic SBOL RDF data
- Part of synbiolib project on sourceforge
- Code
- Documentation
- http://sbol.bhi.washington.edu/py/html/ (help and tell me how put this on sourceforge)
- generated by epydoc http://epydoc.sourceforge.net/manual-usage.html
- tar ball http://sbol.bhi.washington.edu/py/html/sbol3_html.tar.gz
- The API uses and therefore depends on:
- rdflib - for RDF parsing, common operations
- http://www.rdflib.net/2.4.1/
- rdflib is best used under Python 2.5
- pySesame - for access to the Sesame RDF repository and query endpoint
- rdflib - for RDF parsing, common operations
- Also available on:
- the machine running the endpoint
- http://sbol.bhi.washington.edu/py/sbol3.py depends on:
- the machine running the endpoint
Usage
Using the simple web interface
- Find Parts by Search Term or ID and display as a HTML table with or without annotations:
http://sbol.bhi.washington.edu/py/tables_html.py
- Find Parts by ID/ name display in text boxes:
This simple web form demonstrates the use of the getParts() function to retrieve Parts and to print the information it holds as text.
http://sbol.bhi.washington.edu/py/form.py
- Find Parts by Search Term or ID and display as a tab delim table:
This web form allows the user to search the SBOL Semantic repository for a search term within the short and long descriptions.
http://sbol.bhi.washington.edu/py/tables.py
Using the API
Install Python 2.5, rdflib, and then download the sbol3.py and pySesame.py from the sourceforge page
The prototype API offers one main function intended for use. getParts() expects a list of part names (as strings) to find within the repository, it prints a text based version of the part information to standard out, and then returns a list of sbol3.Part objects it found.
parts = sbol3.getParts("BBa_B0010", "BBa_B0012")
You can access the attributes of the parts by:
part1_name = parts[0].name
part2_name = parts[1].name
part1_feature1 = parts[1].annotation[0].feature[0].name
See documentation for other attributes:
Older Versions
Semantic SBOL version v0.1
There is also some data (25MB) represented using Semantic SBOL v0.1(Thanks Emma and Cesar)
For reference: Originally implemented as Provisional BioBrick Language (PoBoL)
Final Version of PoBoL file
######################################################################################################################## ######################################################################################################################## ######################################################################################################################## ########################################################################################################################
Below is an Outdated Work in Progress Section
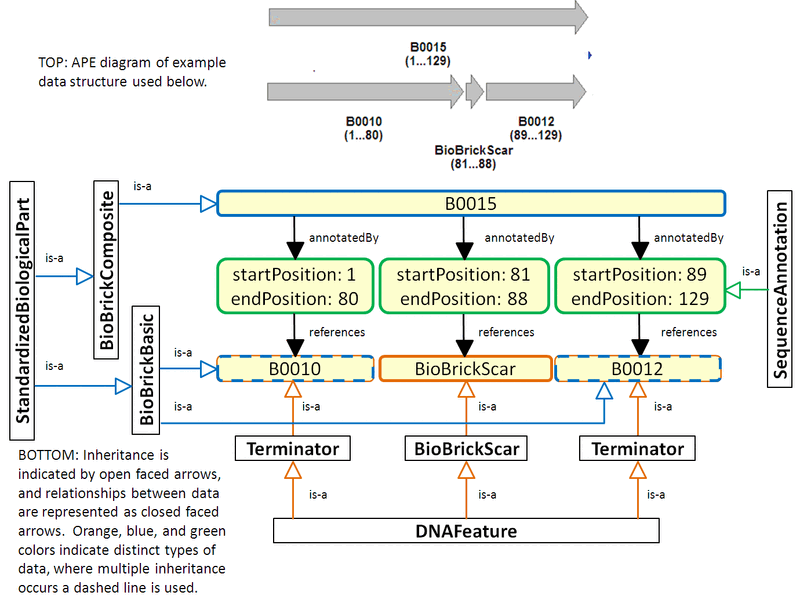
Below is an example based on the composition of BBa_B0015 from B0010 and B0012. It demonstrates the use of Sequence Annotations to annotate the sequence of B0015 with three DNA features: B0010, a BioBrick scar, and B0012. Since B0010 and B0012 are also BioBrick parts, they inherit from both the Standard Biological Part class and the DNA Feature class. The image below is a rough figure demonstrating the description of B0015 in terms of its Parts and Features using Sequence Annotations to specify the position of Features Parts within B0015. As of now, the proposed idea allows Parts to be Features, it does not preclude the specification of subPart relationships as well, see subPart Composition Figure below.
Feature Annotations Figure
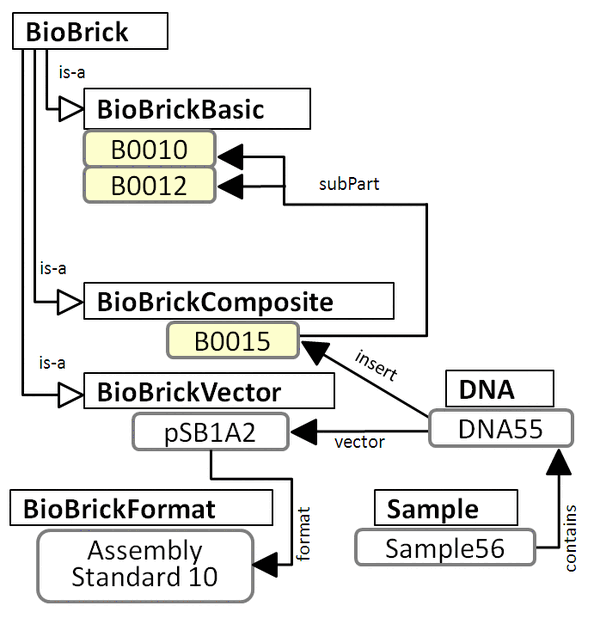
subPart Composition Figure
B0015 Example in OWL format
File:SBOL B0015 AnnotationExample 20090812.owl
SBOL Semantic Reflecting Core Data Model
- The following files are useful for debugging and not in complete sync with the diagrams above, yet...
- This file includes the class definitions and some example individuals of each class.
- The following file represents an example for use in testing queries as it has been processed by a reasoner and therefore includes all the consequences of the semantics. (ie Individuals belong to their asserted types and inferred types)
- sbol_core_debug_example_inferred.owl