The BioBricks Foundation:Standards/Technical/PoBoL
PoBoL ("Provisional BioBrick Language") is the name given to an initial attempt at defining a minimal data model for BioBrick repository entries. It was developed during the Standards and Specifications in Synthetic Biology workshop in Seattle on April 26-27, 2008.
"Pobol" means "people" in Welsh; the developers thought that it was a fitting name for this community-defined format.
Goals
- Provide Pobol ontology and examples (in progress)
- Pobol ontology as RDF/XML
- example Pobol - XML of a Biobrick, Biobrick-Format, Biobrick Family, PhysicalDNA, and Sample record
- clarify where the ontology is hosted (e.g. biobricks.org/rdf)
- Provide validation suite for Pobol - formatted documents (web tool??)
- Implement pobol in freebase.com (Currently in progress)
- emit part entries in pobol
- Implement pobol in BrickIt
- emit part entries in pobol
- absorb part entries in pobol
- Possibly implement pobol in the MIT Registry
- emit part entries in pobol
- demonstrate part exchange between Freebase & Brickit via pobol
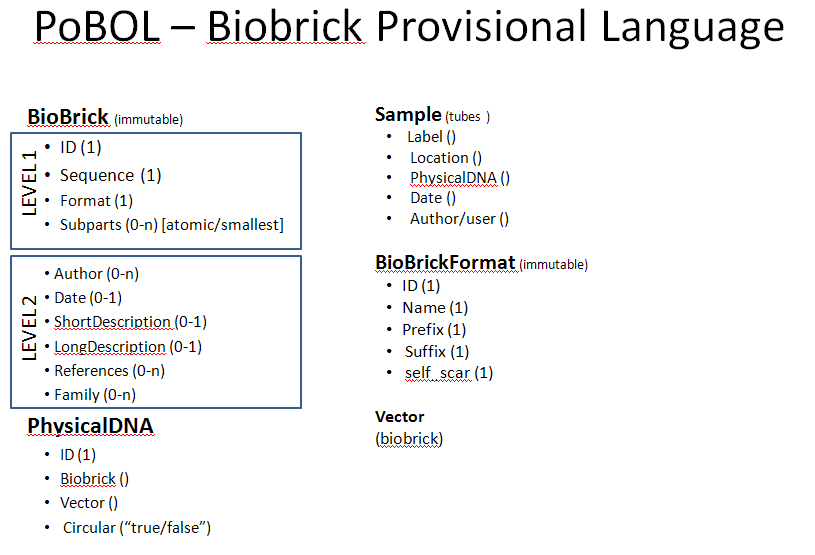
Class Diagrams
OWL / RDF draft
[GoogleCode project for Pobol] The current state of the ontology draft is available from the "Source" repository and will be released regularily.
The draft is available in two standard formats: OWL/RDF and the more human-readable Turtle.The turtle document has so far been polished by hand but we should rather settle on some software tools that help avoiding this.
OWL software recommendations
see also: previous OWW discussion
see also: comprehensive list on W3C wiki
Protege
- Ontology Editor with integrated Triple Store and Reasoner
- imports OWL/XML, exports OWL/XML and (a bit ugly) Turtle
- rather complex user interface
Swoop
- more light-weight ontology editor
- imports OWL/XML, exports OWL/XML
- perhaps easier user interface
- more restricted functionality
- seems to ignore some OWL statements
web rdf validation / translation -- [1]
...add your favorite...
Example: J23100 family
JCA's J23100 promoter family encoded as xml / owl here
Test Cases
The following are suggested test cases for the pobol data model and are derived directly from a discussion on the biobricks technical standards mailing list. As developers of the standard, we have an imperative to not just bake our cake, but to try it too.
Data Entry test cases
- ex: each BB should should have a unique ID, or list of required/optinal fields.
Search test cases
- return all composite sequences having a specific BB
- return all BB being a constitutive promoter in E.Coli - perhaps a test for registries, not for pobol
Data Flow and Data Exchange test cases
- 1 Person / N Parts / 1 Registry
- X Persons / N Parts / 1 Registry
- X Persons / N Parts / M Registries
- Registry 1 wants to import n1 parts from Registry 2
Notes
Feeds
A registry will likely want to expose a feed of its parts. Atom is a good candidate for the feed format (it is an alternative to/improvement upon RSS 2.0 - see Wikipedia for a discussion)
- Atom standard: http://en.wikipedia.org/wiki/Atom_(standard)
- URN:UUID RFC: http://www.ietf.org/rfc/rfc4122.txt
- URN:UUID Provider: http://www.itu.int/ITU-T/asn1/uuid.html