Robert W Arnold Week 9
From OpenWetWare
Jump to navigationJump to search
Electronic Lab Notebook 9
HIV Structure Project
- Started class, determining which sequences to use for each subject, one pre AIDS one post AIDS for first 6 subjects, Subject 6 and 13 first and last visits used
- Subject 1 - visit 1 and 3
- Subject 3 - visit 1 and 5
- Subject 4 - visit 1 and 4
- Subject 10 - visit 1 and 5
- Subject 11 - visit 1 and 4
- Subject 15 - visit 1 and 4
- Subject 6 - visit 1 and 9
- Subject 13 - visit 1 and 5
- Here is the list of sequences and visit data used for our project.
- Hope to determine a V3 conformational change where subjects begin to trend towards AIDS.
In-Class Activity
- Convert your DNA sequences into protein sequences.
- StarBioChem will be used to convert DNA sequences in protein sequence along with NCBI tools.
- The amino acid sequences from Bedrock will be used because they are already there to use, won't have to translate DNA.
- Perform a multiple sequence alignment on the protein sequences.
- Amino acid sequences have fewer differences due to genetic code being redundant. There are multiple ways to code for some amino acids so a simple change of base may not show you any different amino acid or protein sequence. The nucleic acid sequence shows each individual component or the groups of 3 that code for amino acids.
- This is accounted for by redundancy of genetic code as stated above.
- Which of the procedures from Chapter 6 that you ran on the entire gp120 sequence are applicable to the V3 fragment you are working with now?
- In Chapter 6 we were looking for transmembrane proteins using ProtParam. With just the V3 fragment, I think ProtParam will again be useful as it will be able to tell the change in residues between different clones and the composition of the V3 region.
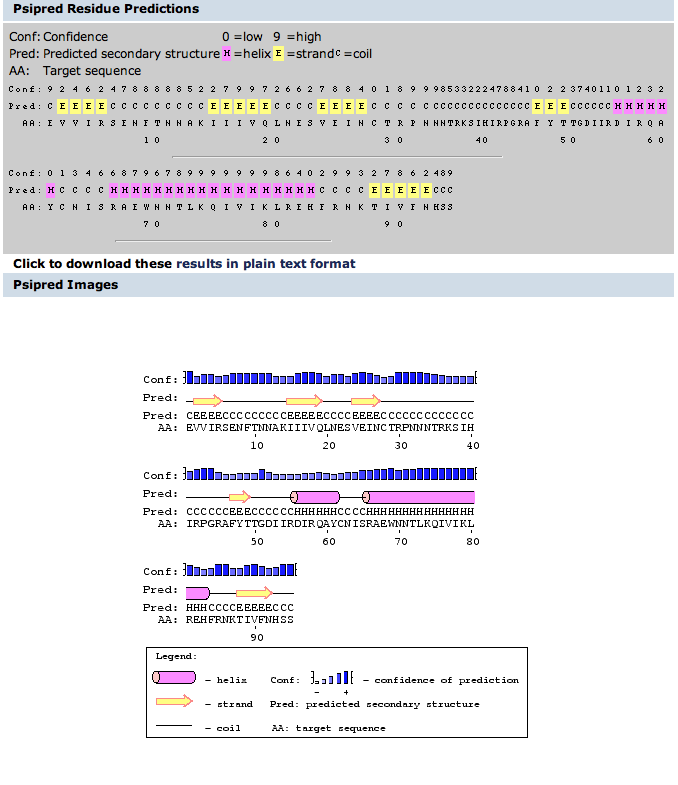
- Chapter 11 contains procedures to use for working with protein 3D structures. Find the section on "Predicting the Secondary Structure of a Protein Sequence" and perform this on both the entire gp120 sequence and on the V3 fragment that we are now working with. You will compare the predictions with the actual structures.
- Opened book to page 330.
- Went to bioinf.cs.ucl.ac.uk/psipred/ and looked for button to click to enter server, it wasn't there.
- Used gp120 sequence from Kwong link below and here were results:
- Download the structure files for the papers we read in journal club from the NCBI Structure Database.
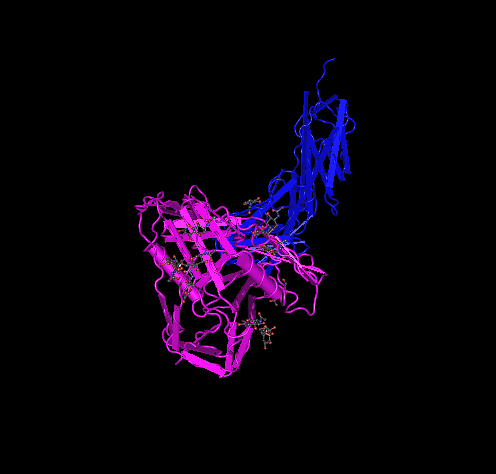
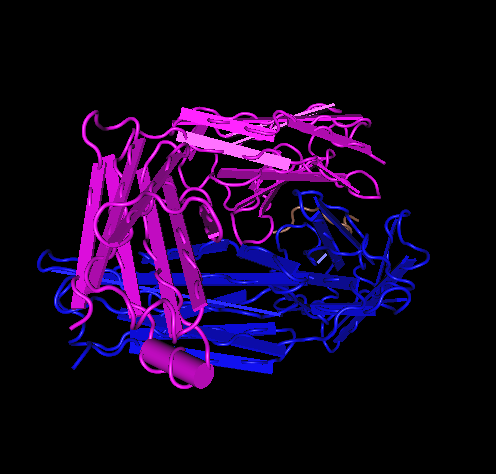
- Here is the Kwong 1GC1 structure:
- Here are the gp120 prediction results from the Kwong paper.
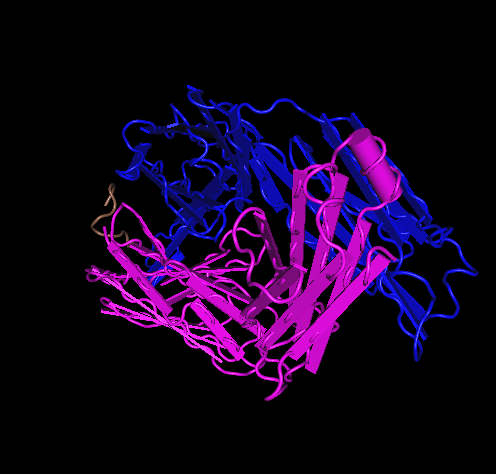
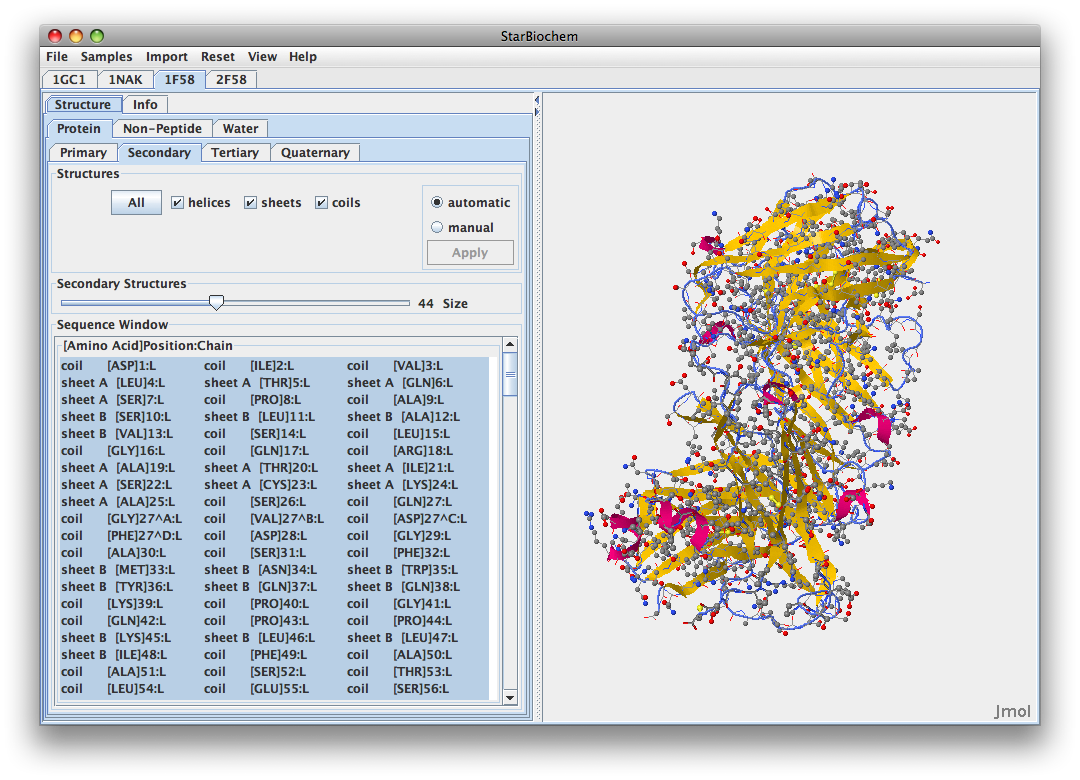
- Here is the Stanfield 1999 1F58 structure:
- Here is the Stanfield 1999 2F58 structure:
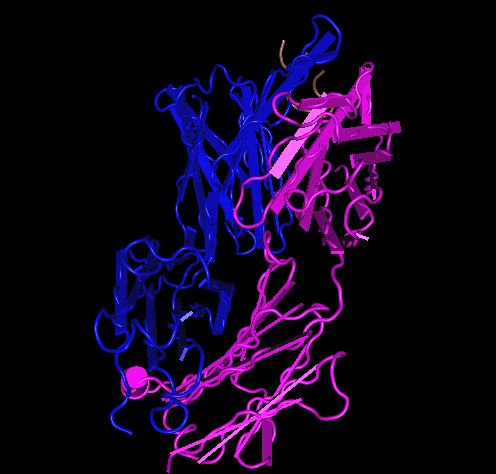
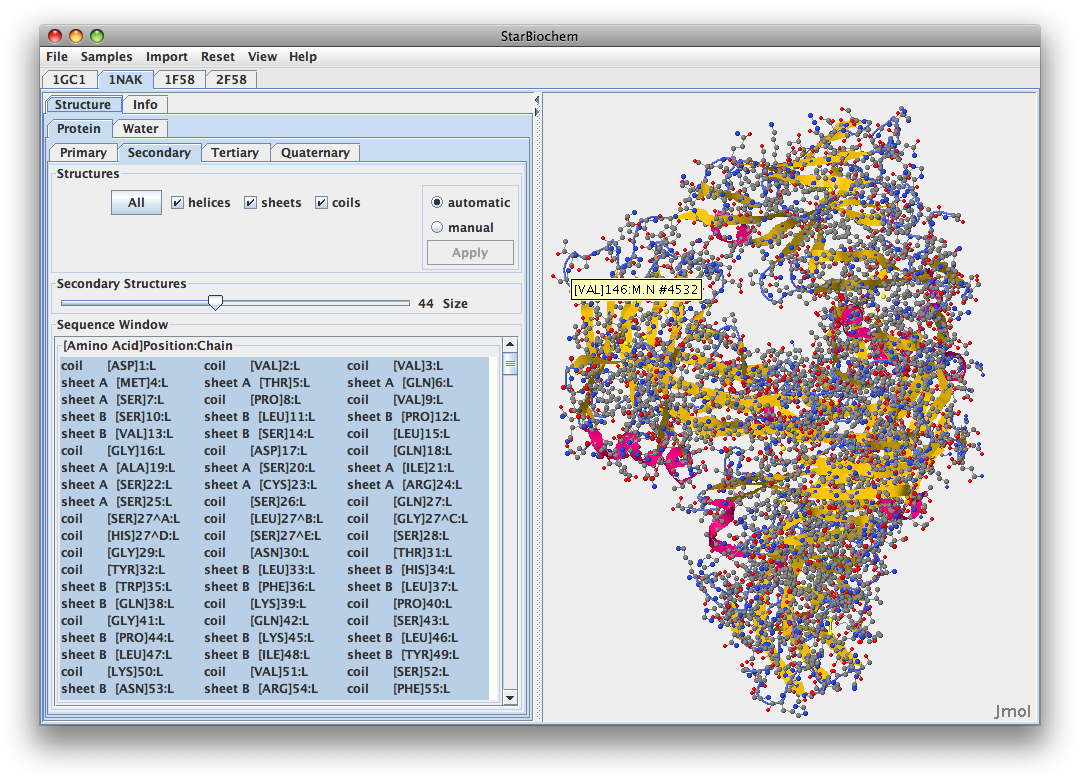
- Here is the Stanfield 2003 1NAK structure:
- These files can be opened with the Cn3D software site that is installed on the computers in the lab (this software is free, so you can download it and use it at home, too.) Familiarize yourself with the software features (rendering and coloring) with both the gp120 peptide and ternary complex structures. Alternately, you may choose to use the Star Biochem program to do this portion of your work. Answer the following:
- Opened up StarBiochem
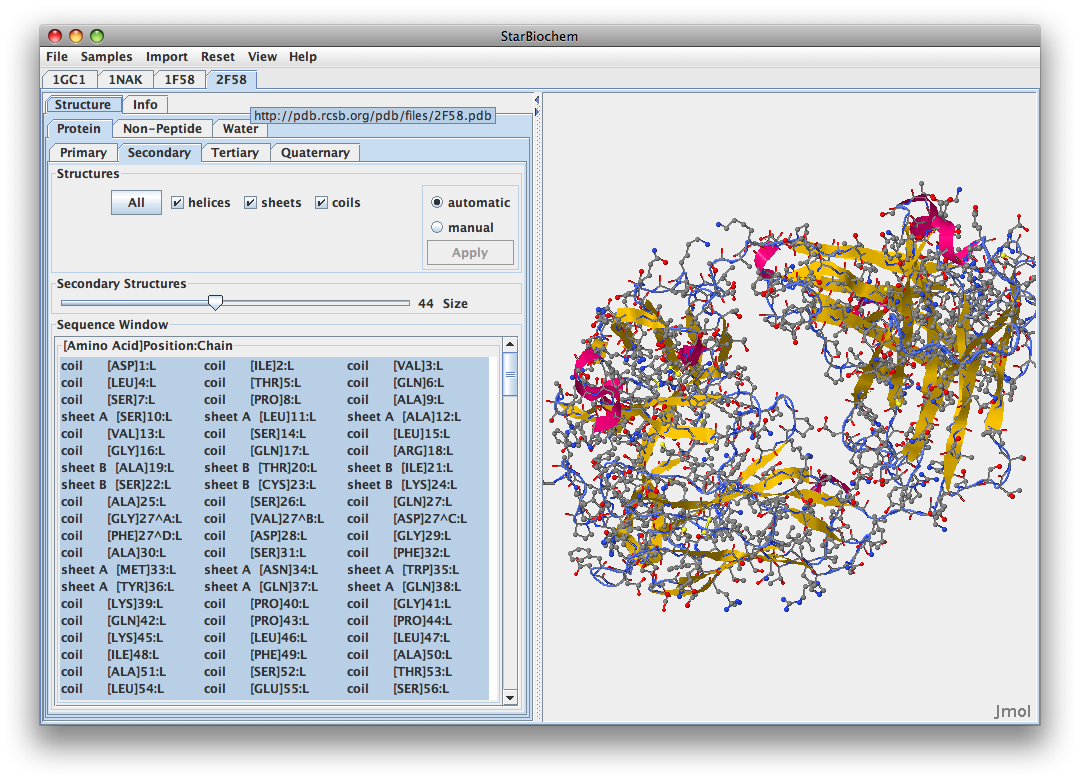
- Imported all structres: 1GC1, 1F58, 2F58, 1NAK.
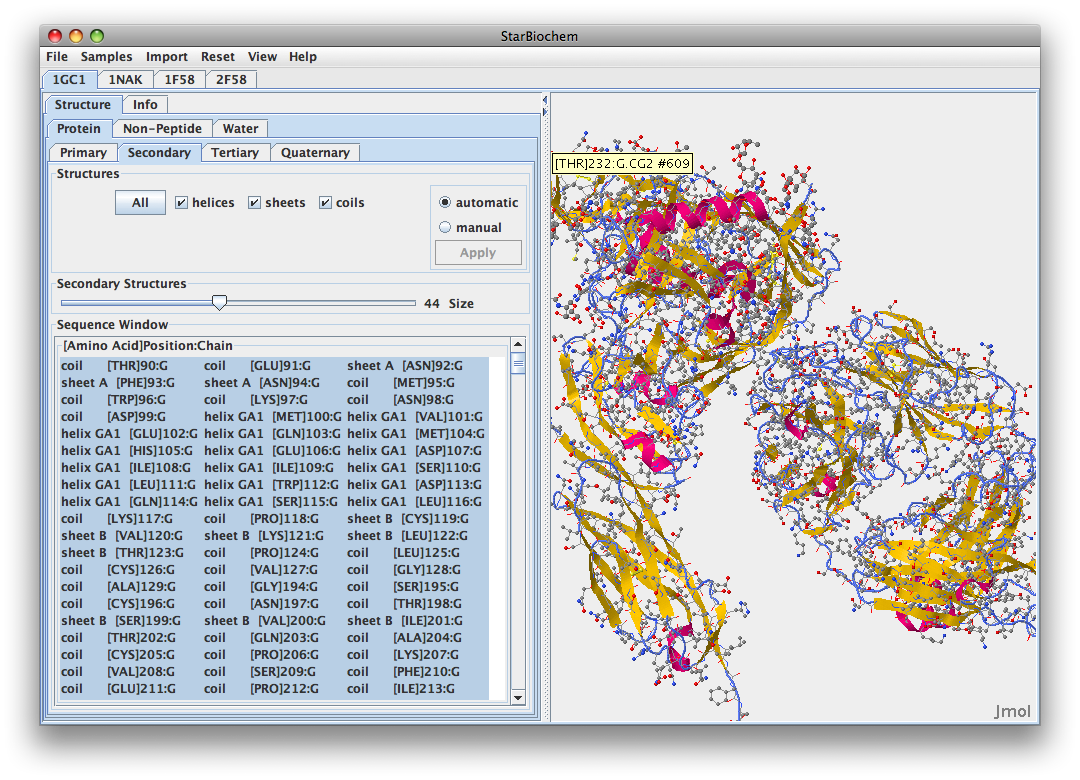
- 1GC1
- 1F58
- 2F58
- 1NAK
- 1GC1 is composed of three peptides. gp120 G has its N-terminus at THR position 90 and C-terminus at GLU position 492. gp120 domain C has N-terminus at LYS position 1 and C-terminus at LYS at position 181. The antibody has N-terminus at GLU position 1 and C-terminus at ARG position 213.
- 1F58 has its N at ASP position 1 and C-terminus at THR position108
- 1NAK has its C-terminus at GLU position 204
- PSIPRED worked well, but I wasn't exactly sure how to predict secondary structures and some residues I felt did not match up. For the most part though, it looked how I thought it was going to.
- V3 region from Kwong 1GC1 is not there because it was cut out for crystallization of the gp120. In between the blue coils and yellow sheet in the picture would be the location of the V3.
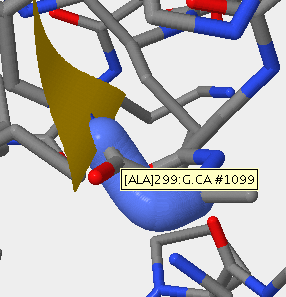
- V3 1F58 313P-325P.
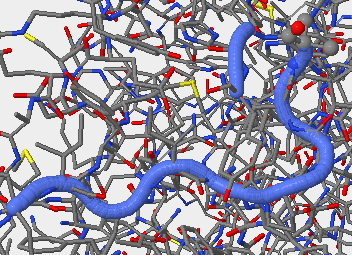
- V3 2F58 315P-324P.
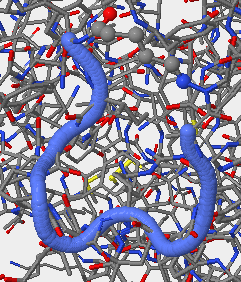
- V3 1NAK 312P-323P.
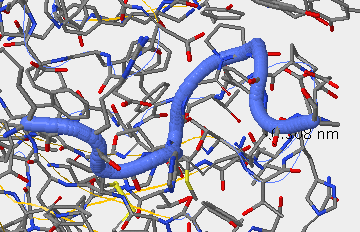
- Once you have oriented yourself, analyze whether the amino acid changes that you see in the multiple sequence alignment would affect the 3D structure and explain why you think this.
- Here are the results from the multiple sequence alignments from the subjects visits pre and post AIDS:
#*Subject 1 Before and After AIDS Alignment
S1V5-4 EVVIRSENITNNAKIIIVQLNESVAISCTRPNNNIKQRIMHIRPGRAFYTK-DITEDIRQ
S1V1-2 EVVIRSENFTNNAKIIIVQLNESVEINCTRPNNNTR-KSIHIRPGRAFYTTGDIIRDIRQ
********:*************** *.******* : : :**********. ** .****
S1V5-4 AYCNISRTAWNNTLKQIVKKLREHFVNKTIVFNHSS
S1V1-2 AYCNISRAEWNNTLKQIVIKLREHFRNKTIVFNHSS
*******: ********* ****** **********
#*Subject 3 Before and After AIDS Alignment
S3V6-6 DIVIRSANFSDNAKTILVQLNETVVMNCTRPGNNTRKRVTLGPGRVYYTTGQIIGDIRKA
S3V1-3 DVVIRSANFTNNAKTILVQLNETVVMNCTRPGNNTRKRVTLGPGKVYYTTGQIIGDIRKA
*:*******::*********************************:***************
S3V6-6 HCNLSRAGWNSTLERIAIKLREQFQNKTIAFNQSS
S3V1-3 HCNLSRADWNNTLKRIAIKLREQFQNKTIAFNQSS
*******.**.**:*********************
#*Subject 4 Before and After AIDS Alignment
S4V4-2 EVVIRSENFTNNAKIIIVQLNESVEINCTRPDNHTVRKIPIGPGRSFYTTGIVGDIRQAH
S4V1-1 EVVIRSENFTNNAKIIIVQLNKSVEINCTRPNNNTIRRIPIGPGRAFYTTGRIGDIRPAH
*********************:*********:*:*:*:*******:***** :**** **
S4V4-2 CNISKTKWNNTLKLIVNKLREQFGNKTIIFNQSS
S4V1-1 CNISRTKWNNTLKLIVNKLREQFRNKTIIFNQSS
****:****************** **********
#*Subject 10 Before and After AIDS Alignment
S10V6-1 EVVIRSENFTDNAKTIIVHLNKSVEINCTRPNNNTRRSINMGPGRAFYATGEIIGDIRQA
S10V1-7 EVVIRSENFTDNAKTIIVQLNKSVEINCTRPNNNTRRSINMGPGRAFYTTGEIIGDIRQA
******************:*****************************:***********
S10V6-1 HCNLSRTKWNDTLKQVVAKLREQFRNKTIIFTQSS
S10V1-7 HCNLSRTKWNDTLKQVVDKLGEQFRNKTIIFNQSS
***************** ** **********.***
#*Subject 11 Before and After AIDS Alignment
S11V4-5 EVIIRSENFSNNAKNIIVQLNESVVINCTRPDNTIKQRIIHIGPGRPFYTTGIKGNIRQA
S11V1-3[4] EVIIRSENFSNNAKNIIVQLNESVVINCTRPDNTIKQRIIHIGPGRPFYTTGIKGNIRQA
************************************************************
S11V4-5 HCNVSRGQWNKTLEQVVRKLREQYGPNKTIVFKQPI
S11V1-3[4] HCNVSRGQWNKTLEQVVRKLREQYGLNKTIVFKQPI
************************* **********
#*Subject 15 Before and After AIDS Alignment
S15V4-7 EVVIRSENFTNNAKIIIVHLNESVVINCTRPNNNTRRKIHIGPGKTFYTGDIIGNIRQAH
S15V1-10 GVVIRSENFTNNAKIIIVQLKEAVRINCIRPNNNTRRRIPIGPGSAFYTTGIIGDIRQAH
*****************:*:*:* *** ********:* ****.:*** .***:*****
S15V4-7 CNISGSKWNNTLKQIVNKLREQFGNKTIVFNQSS
S15V1-10 CNISGSKWNSTLKQIVNKLREQFVNKPIIFNQSS
*********.************* **.*:*****
#*All Rapid Progressor Subjects Post-AIDS Alignment
S11V4-5 EVIIRSENFSNNAKNIIVQLNESVVINCTRPDNTIKQRIIHIGPGRPFYTT-GIKGNIRQ
S1V5-4 EVVIRSENITNNAKIIIVQLNESVAISCTRPNNNIKQRIMHIRPGRAFYTK-DITEDIRQ
S15V4-7 EVVIRSENFTNNAKIIIVHLNESVVINCTRPNNNTR-RKIHIGPGKTFYTG-DIIGNIRQ
S4V4-2 EVVIRSENFTNNAKIIIVQLNESVEINCTRPDNHT-VRKIPIGPGRSFYTT-GIVGDIRQ
S10V6-1 EVVIRSENFTDNAKTIIVHLNKSVEINCTRPNNNTR-RSINMGPGRAFYATGEIIGDIRQ
S3V6-6 DIVIRSANFSDNAKTILVQLNETVVMNCTRPGNNTR-KRVTLGPGRVYYTTGQIIGDIRK
:::*** *:::*** *:*:**::* :.****.* : : : **: :*: * :**:
S11V4-5 AHCNVSRGQWNKTLEQVVRKLREQYGPNKTIVFKQPI
S1V5-4 AYCNISRTAWNNTLKQIVKKLREHF-VNKTIVFNHSS
S15V4-7 AHCNISGSKWNNTLKQIVNKLREQFG-NKTIVFNQSS
S4V4-2 AHCNISKTKWNNTLKLIVNKLREQFG-NKTIIFNQSS
S10V6-1 AHCNLSRTKWNDTLKQVVAKLREQFR-NKTIIFTQSS
S3V6-6 AHCNLSRAGWNSTLERIAIKLREQFQ-NKTIAFNQSS
*:**:* **.**: :. ****:: **** *.:.
#*Control Subjects 6 & 13 Alignment
S13V5-1[4] EIVIRSENFTNNAKIIIVQLKESVEINCTRPGNNTRRSINIGPGRAFYASRGIIGDIRQA
S6V9-3 EVVIRSANLTDNAKIIIVHLNESVEMNCTRPNNNTRKGIHIGPGRAFYATGEIIGNIRQA
*:**** *:*:*******:*:****:*****.****:.*:*********: ***:****
S13V5-1[4] YCNISKAKWDNTLGQVAAKLREQFRNATIVFNQSS
S6V9-3 HCNLSRAPWNDTLKRIAIKLREQFKNKTIAFNQSS
:**:*:* *::** ::* ******:* **.*****
#*Alignment of Control Subject 13 Sequence to Post-AIDS Sequences from all Subjects
S13V5-1[4] EIVIRSENFTNNAKIIIVQLKESVEINCTRPGNNTR-RSINIGPGRAFYASRGIIGDIRQ
S15V4-7 EVVIRSENFTNNAKIIIVHLNESVVINCTRPNNNTR-RKIHIGPGKTFYTG-DIIGNIRQ
S1V5-4 EVVIRSENITNNAKIIIVQLNESVAISCTRPNNNIKQRIMHIRPGRAFYTK-DITEDIRQ
S10V6-1 EVVIRSENFTDNAKTIIVHLNKSVEINCTRPNNNTR-RSINMGPGRAFYATGEIIGDIRQ
S4V4-2 EVVIRSENFTNNAKIIIVQLNESVEINCTRPDNHTV-RKIPIGPGRSFYTT-GIVGDIRQ
S11V4-5 EVIIRSENFSNNAKNIIVQLNESVVINCTRPDNTIKQRIIHIGPGRPFYTT-GIKGNIRQ
S3V6-6 DIVIRSANFSDNAKTILVQLNETVVMNCTRPGNNTR-KRVTLGPGRVYYTTGQIIGDIRK
:::*** *:::*** *:*:*:::* :.****.* : : : **: :*: * :**:
S13V5-1[4] AYCNISKAKWDNTLGQVAAKLREQFR-NATIVFNQSS
S15V4-7 AHCNISGSKWNNTLKQIVNKLREQFG-NKTIVFNQSS
S1V5-4 AYCNISRTAWNNTLKQIVKKLREHFV-NKTIVFNHSS
S10V6-1 AHCNLSRTKWNDTLKQVVAKLREQFR-NKTIIFTQSS
S4V4-2 AHCNISKTKWNNTLKLIVNKLREQFG-NKTIIFNQSS
S11V4-5 AHCNVSRGQWNKTLEQVVRKLREQYGPNKTIVFKQPI
S3V6-6 AHCNLSRAGWNSTLERIAIKLREQFQ-NKTIAFNQSS
*:**:* *:.** :. ****:: * ** *.:.
#*Alignment of Control Subject 6 Sequence to Post-AIDS Sequences from all Subjects
S6V9-3 EVVIRSANLTDNAKIIIVHLNESVEMNCTRPNN-NTRKGIHIGPGRAFYATGEIIGNIRQ
S11V4-5 EVIIRSENFSNNAKNIIVQLNESVVINCTRPDNTIKQRIIHIGPGRPFYTT-GIKGNIRQ
S1V5-4 EVVIRSENITNNAKIIIVQLNESVAISCTRPNNNIKQRIMHIRPGRAFYTK-DITEDIRQ
S4V4-2 EVVIRSENFTNNAKIIIVQLNESVEINCTRPDN-HTVRKIPIGPGRSFYTT-GIVGDIRQ
S15V4-7 EVVIRSENFTNNAKIIIVHLNESVVINCTRPNN-NTRRKIHIGPGKTFYTG-DIIGNIRQ
S10V6-1 EVVIRSENFTDNAKTIIVHLNKSVEINCTRPNN-NTRRSINMGPGRAFYATGEIIGDIRQ
S3V6-6 DIVIRSANFSDNAKTILVQLNETVVMNCTRPGN-NTRKRVTLGPGRVYYTTGQIIGDIRK
:::*** *:::*** *:*:**::* :.****.* . : : : **: :*: * :**:
S6V9-3 AHCNLSRAPWNDTLKRIAIKLREQFK-NKTIAFNQSS
S11V4-5 AHCNVSRGQWNKTLEQVVRKLREQYGPNKTIVFKQPI
S1V5-4 AYCNISRTAWNNTLKQIVKKLREHF-VNKTIVFNHSS
S4V4-2 AHCNISKTKWNNTLKLIVNKLREQFG-NKTIIFNQSS
S15V4-7 AHCNISGSKWNNTLKQIVNKLREQFG-NKTIVFNQSS
S10V6-1 AHCNLSRTKWNDTLKQVVAKLREQFR-NKTIIFTQSS
S3V6-6 AHCNLSRAGWNSTLERIAIKLREQFQ-NKTIAFNQSS
*:**:* **.**: :. ****:: **** *.:.
- The article we chose is "Indentification and structural characterization of novel genetic elements in the HIV-1 V3 loop regulation coreceptor usage" by Valentina Svicher et al.
- The article cannot be uploaded on to this site due to copyright.
- The background of this article dealt with the interaction between HIV-1 gp120 and the CCR5 terminus. Svicher is trying to study how the different genetic signatures of gp120 V3 domain effect the interaction strength.
- 251 patients were used in study and HIV-1 coreceptor usage and V3 sequencing were tested. Gp120 interaction with CCR5 terminus was also evaluated.
- Results shows that selected V3 correlate significantly with CCR5 or CXCR4 usage and modulate gp120 affinity for CCR5 terminus.
- Found "key determinants for tropism within the V3 sequence, confirmed by structure- and by phenotypic-tropism." Going to potentially be used to find how to manipulate CCR5 and to "provide molecular implications for design of new entry inhibitors."
Updated ppt with work from Alex and Zeb
Links
- Robert W Arnold Week 2
- Robert W Arnold Week 3
- Robert W Arnold Week 4
- Robert W Arnold Week 5
- Robert W Arnold Week 6
- Robert W Arnold Week 7
- Robert W Arnold Week 8
- Robert W Arnold Week 9
- Robert W Arnold Week 10
- Robert W Arnold Week 11
- Robert W Arnold Week 12
- Robert W Arnold Week 14
- Week 2 Assignment
- Week 3 Assignment
- Week 4 Assignment
- Week 5 Assignment
- Week 6 Assignment
- Week 7 Assignment
- Week 8 Assignment
- Week 9 Assignment
- Week 10 Assignment
- Week 11 Assignment
- Week 12 Assignment
- Week 14 Assignment
- Class Journal Week 1
- Class Journal Week 2
- Class Journal Week 3
- Class Journal Week 4
- Class Journal Week 5
- Class Journal Week 6
- Class Journal Week 7
- Class Journal Week 8
- Class Journal Week 9
- Class Journal Week 10
- Class Journal Week 11
- Class Journal Week 12
- Class Journal Week 14