Restriction digest
General Information
Restriction digest involves the cutting of DNA at specific recognition sequences by enzymes.
General Procedure
Specific Protocols
Enzyme selection for BioBricks digest
Richard Lab:Restriction Digest
Restriction Ligation in Genome Compiler
An intuitive cloning wizard for simulating digest and ligate with restriction enzymes or with PCR can be found in Genome Compiler
How does it work?
Open a new construction project and select the restriction ligation method. Start with the Insert tab within the cloning wizard:
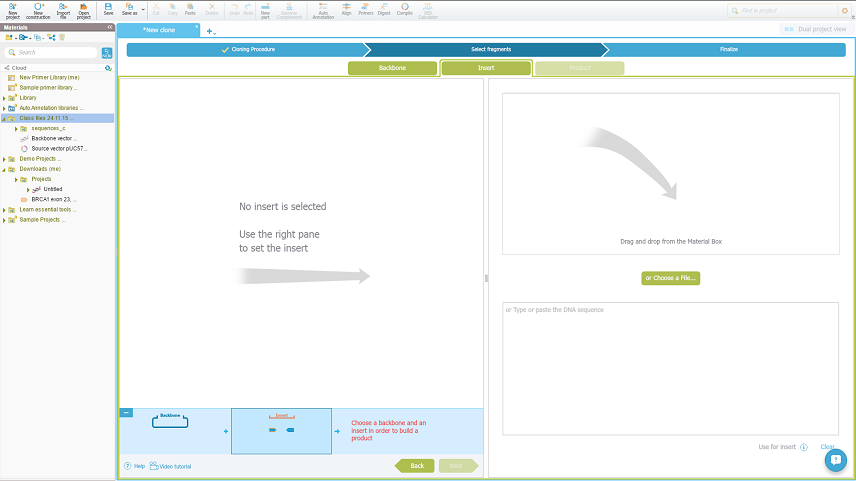
Than just drag and drop your source vector with the gene of interest, select to generate by digest, choose the restriction enzymes out from the lists and choose the the insert fragment:
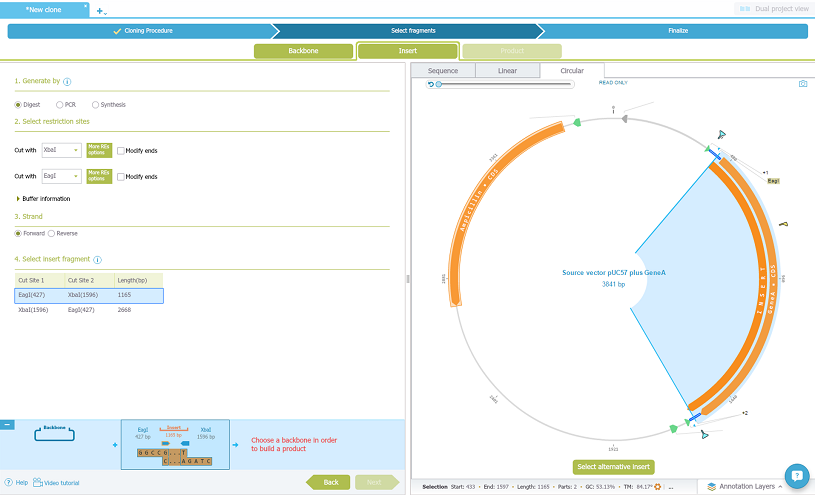
Now select the Backbone tab. Drag and drop the backbone vector from the materials box on the left, choose to linearise it by digest and choose the restriction enzymes, and select the fragment:
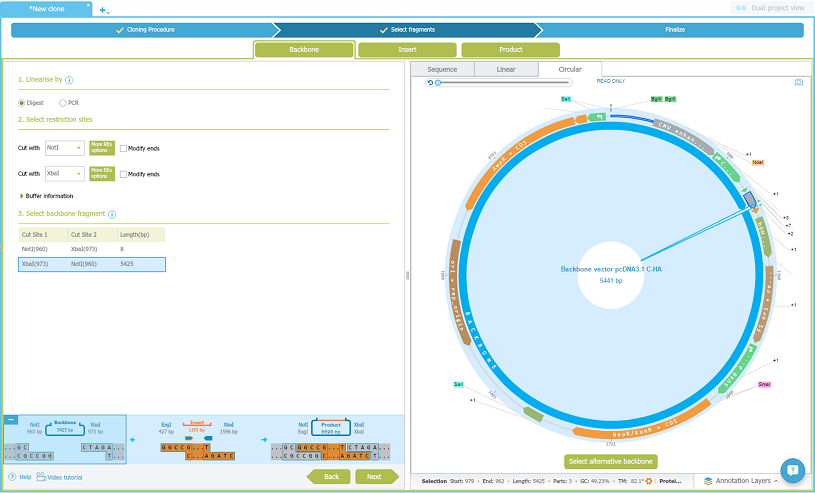
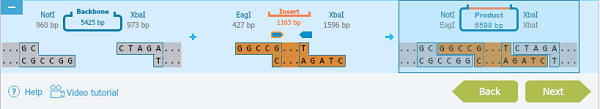
Check the final product:
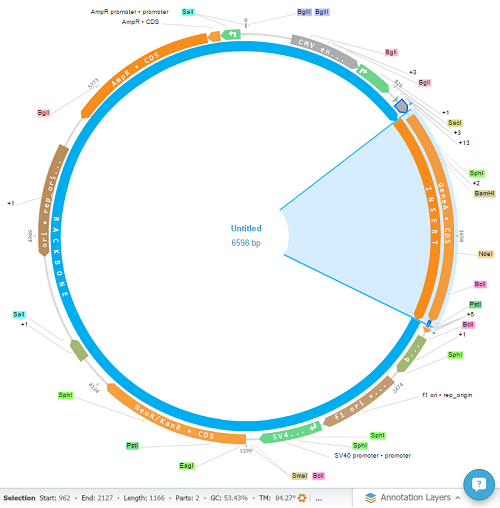
And view the cloning process summary:
Notes
- For help with specific enzymes, see Restriction enzymes
- For buffer composition, see Restriction digest/Buffers
- If you are interested in cutting near the ends of the linear DNA fragment, note that some enzymes do not cut efficiently at the ends of linear DNA. So include extra bases to increase the efficiency of cutting. Many enzymes work with 4 bases supposedly but XhoI was found to require more than 4 bases (8 bases was used successfully). Thus, to be on the safe side, use 8 bases whenever possible. NEB has more information here. Read the information at NEB carefully ... they recommend adding 4 bases to the numbers listed in their table.
- Tom was once having some trouble with failed restriction digests. He called NEB and they recommended doing the digest in a larger volume. Therefore, the Knight lab typically does digests in either 50 μL or 100 μL volumes rather than the 20 μL volume that the Endy lab uses. If the sample needs to be concentrated, some method of DNA purification is used.