NanoBio: Integration into yeast
From OpenWetWare
Jump to navigationJump to search
Back to NanoBio
Back to Protocols
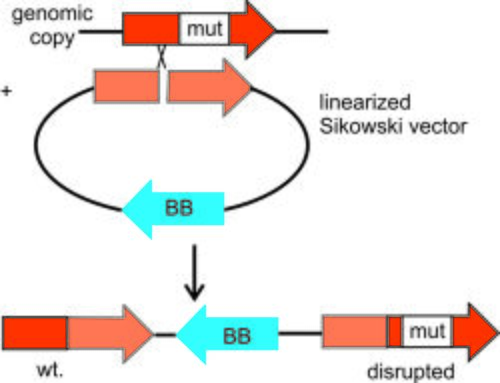
Background
Materials
- 5X TE/LiOAc
- 2.5 mL 1M LiOAc
- 0.25 mL 1M Tris-HCl pH 8.0
- 0.1 mL 0.5M EDTA
- 2.15 mL ddH2O
- 40% PEG / 1X TE / LiOAc
- 1 mL 5X TE/LiOAc
- 4 mL 50% PEG
- DMSO
- Single-stranded carrier DNA
Protocol
Day 0
- Inoculate the yeast strain into 5 ml of liquid medium and incubate overnight.
- If integrating into the genome, linearize at least 3.75 ug plasmid by digestion with the appropriate enzyme. This enzyme should cut the vector singly such that at one of the ends of the resulting linearized DNA, the wild type allele has ~100 bp overlap with the auxotrophic allele.
- PRS303 (HIS3) use PstI
- pRS304* (modified TRP1) use PstI
- pRS305 (LEU2) use BstEII
- pRS306 (URA3) use PstI
- PstI digestions should be performed overnight at 37C, while BstEII digestions should be performed for ~ 4 hours at 37C.
Day 1
- Measure the OD600 and dilute it until you get to ~0.1 OD. For example, if you have an OD600 of 5, you want dilute it 50 times.
- Grow it until you achieve an OD600 of 0.4-0.7. (You should at least have 2 doublings before transforming)
- If integrating, check that the plasmid is linearized, but not fragmented by multiple cuts. Do this by running ~1 uL of the digest and a similar amount of uncut plasmid on an agarose gel. The digested plasmid can be used without further purification or can be purified using a Qiagen pcr cleanup kit or ethanol precipitation. I (Caroline) prefer to pcr purify the digest, using 20 uL to elute the DNA. In any case, it is important that the concentration of linearized DNA is at least 250 ng/uL. (tip: run a gel w/ 1 uL to get a feeling of how much ng/uL you have before pcr purification if you want to do it, BA)
- Tranfer the cells to falcon tubes and centrifuge for 4min at 2300rpm. Decant.
- Re-suspend the cells in 10 mL ddH2O. Centrifuge for 4min at 2300rpm. Decant.
- Re-suspend the cells in 1 mL ddH2O into eppendorf tubes.
- Centrifuge for 10 seconds at 14000 rpm. Decant.
- Re-suspend in 1mL 1X TE/LiOAc buffer. Centrifuge for 10 seconds at 14000 rpm and decant. Repeat.
- Re-suspend in 50µL 1X TE/LiOAc buffer. Let sit @ 4°C until ready
- While waiting, boil ssDNA for 5min then let cool to room temperature
- Add 4µL of the digested MPs to each 50µL of cells plus 2µL ssDNA.
- Incubate for 35min at 30°C
- Add 300µL 40% PEG in 1X TE/LiOAc. Vortex well.
- Incubate for 35min at 30°C in eppendorf inverter
- Add 35µL DMSO
- Heat-shock at 42°C for 15min.
- Centrifuge the cells for 10 seconds at 14000 rpm and re-suspend in 150µL ddH2O
- Add cells to appropriate plates according to your DNA and strain
- Put plates at 30°C
Wait a couple days before checking if you have colonies.