Li Lab
Please visit our new lab website https://sites.uci.edu/weililabComputational Cancer Epigenomics and Transcriptomics (PI: Wei Li)Division of Computational Biomedicine, Department of Biological Chemistry, School of Medicine, University of California, Irvine |
Recent News. 03/2021: Yipeng's single cell APA paper was accepted to Genome Research . 02/2021: Lei's 3'aQTL paper entitled "An atlas of alternative polyadenylation quantitative trait loci contributing to complex trait and disease heritability" was accepted to Nature Genetics . 09/2020: Jie's DORGE cancer driver gene paper was accepted to Science Advances . 08/2020: Jianfeng's CHALM methylation paper was accepted to Nature Communications . 08/2020: Cui Ya's NG-Circos paper was accepted to NAR Genomics and Bioinformatics . 01/2020: Our lab has moved to University of California Irvine . 01/2020: Lei's paper w/ Ryan Potts was accepted to Molecular Cell . 10/2019: Collaborative work with David Allis and Wen Hong was accepted to Nature; . 10/2019: Collaborative work with Suming Huang was accepted to Cancer Cell; . 9/2019: Collaborative work with Costa-Mattioli's group was accepted to Science; . 05/2019: Collaborative work with Paolo Sassone-Corsi at UCI was published in Cell . 02/2019: Our new 5-year NCI R01 grant regarding DNA methylation canyon was funded. . 07/2018: Jianzhong's DNA methylation canyon paper was accepted to Genome Biology. . 06/2018: Jiejun’s paper was accepted to Nature Cell Biology. . 06/2018: Xueqiu's DNMT3A binding paper was accepted to Genome Biology . 03/2018: HJ Park's papers were accepted to PLOS Computational Biology and Cell Reports, respectively. . 01/2018: HJ Park's 3'UTR vs ceRNA paper was finally accepted to Nature Genetics after 1.5 years and 3 rounds of revisions/appeals. A good start to the new year!
| |
|
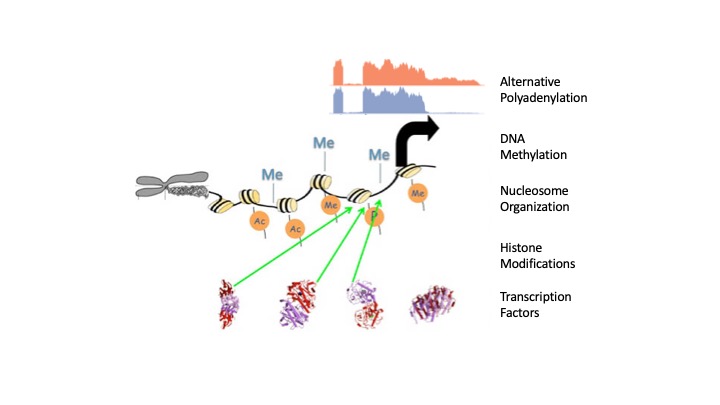
Our lab is focused on the design and application of bioinformatics algorithms to elucidate global regulatory mechanisms in normal development and diseases such as cancer. We have a solid track record in large-scale genomics data analysis, and in developing widely used open-source bioinformatics software, such as MACS (>7,000 citations) for ChIP-seq and RSeqQC (>800 citations) for RNA-seq. In collaboration with experimental biologists, we have used bioinformatics to gain novel biological insights into development, aging, stem cell, neurologic disorders, and various cancers. Our areas of expertise include 1) DNA methylation using Bisulfite-seq; 2) Epigenetic regulation using ChIP-seq; 3) Alternative Polyadenylation (APA); 4) Non-coding RNA; 5) Nucleosome organization using MNase-seq. Since establishing our own bioinformatics lab in early 2008, we have (as of Febuary 2020)
|