Lab Notebook - Gabriela
June
Tuesday, June 16
TECAN Assay Round 1:
1) Read OD600 of cells to determine cell concentration:
Column 1, Rows ABC: positive control (Terry pipette up/down, may have sheared cells) Column 2, Rows ABC: negative control
Readings: A1: 0.2492 A2: 0.3188 B1: 0.2693 B2: 0.3519 C1: 0.2040 C2: 0.2893 D1: 0.0398 D2: 0.0421 E1: 0.0340 E2: 0.0357 F1: 0.0350 F2: 0.0365
D-F are blanks
2)Setup TECAN plate as follows: ABC, 4-7: 25uL liquid cell culture, 75 uL PBS DEF, 4-7: 40uL liquid cell culture, 60 uL PBS columns 4 and 5 have positive control cells (arabinose-induced pBca9494ca-M10220;add .5 uL of streptavidin-R-phycoerythrin conjugate columns 6 and 7 have negative control cells (uninduced pBca9494ca-M10220); add no strept
- Take note to try diluting strept in PBS before adding to cells next time as it is a bit sticky.
5:05 pm incubate at 37 degrees C 5:35 pm remove plate from TECAN
Spin plate 5,400 rpm 5 min.
View under Microscope: .1 second exposure images taken and saved on computer on lab bench in B144.
Wednesday, June 17
Took note results of TECAN experiment (6/16):
- Cells at midlog don't pellet- they form a monolayer
- Try using more saturated cells next time
- TECAN results from 140 class were opposite expected results, possibly due to background fluorescence of pelleted cells.
- On the microscope, focus is an issue
Suggestions (made by Terry):
- use saturated cells
- add 95 ul cells + 5 ul strept in PBS
Come up with possible answers to: 1) How can we make TECAN work? 2) How can we make the microscope work? 3) Can we just spin the plate down and image the whole plate on a UV box? 4) Should we be measuring OD600 before any of this?
Made glucose LB media solution:
- Stock glucose 20% is in wight by volume (g/ml)
- eg. 100 ml 20% -> 20 g glucose / 100 ml water -> filter sterilize -> 2.5 ml solution into 500 ml media(LB)
OD Measurements:
A600 Blank: 1 ml PBS Sample #1 (colony 1) pBca 9495-M10220 diluted 250 ul cells + 750 ul PBS
- A600 = 0.170
- concentration = 8.52e7 cell/ml
Sample #2 (colony 2) pBca 9495-M10220
- A600 = 0.134
- concentration = 6.69e7 cells/ml
Sample #3 (saturated colony 1 M10220)
- A600 = 0.796
- concentration = 3.98e8 cells/ml
Sample #4 (saturated colony 2 M10220)
- A600 = 0.780
- concentration = 3.90e8 cells/ml
Thursday, June 18
- Today we took a closer look at the TECAN assay and program.
- Want to know what the meaning of z-position?
- Range is 1220 to 13000 um
- 1220 um readjusted during a test run to 7220 um as a min.
- 13000 um resulted in readings labeled OVER ->readings possibly too high?
- Readings indicate that lower level z-positions correspond to higher levels in the fluid.
Friday, June 19
- Oligos arrived today
- Set up PCA reaction:
- When oligos arrive take tape off, spin (balance with tray full of water) 5000 rpm 2 min,
- Set up PCA in Biomek program
- Biomek 3000 (file -> oligo plate distributor)
- Set up PCA reaction:
- Continue working on TECAN assay: decide to come in on Sunday to inoculate
Monday, June 22
- Run analytical gels for 12 parts that were synthesized (don't do digestion yet)
- Parts are: ig 1, 27, 57, 79, 113, 121, 129, 137, 145, 171, 201, 207
- Bands visible in all wells
- Digestion of PCR products protocol used for digestion of 12 parts
- Cocktail mixture for 13 x ( 13 ul NEBR, 6.5 ul EcoR1, 6.5 ul BamH1)
- Vector digest completed by Patrick and Susan
- Gel purification (1):
- 4 ul of ladder, 2 ul of loading buffer + 10 ul digestion
- Gel 1 (left to right): ig 1, 27, 57, 79, 113, 121, vector, vector, ladder
- Gel 2 (left to right): vector, vector, 129, 137, 145, 171, 201, 207, ladder
- Need to re-digest 129, 137, 201, and 207 (bands ran off the gel)
- Gel purification (2):
- Lanes (left to right): vector, vector, vector, 207, 201, 137, 129, ladder
- Gel zymo 207, 201, 137, and 129 (need to add isopropanol before spinning <300bp length)
- Ligation: followed normal protocol, except added 2 ul of insert for ig79
- Transformation: added 70 ul of cell cocktail instead of 75 ul
- Plating: added 75 ul of cells (stored in upstairs incubator overnight)
Tuesday, June 23
Check plates: at 10:15 am sparse, small colonies; will let them incubate for a little longer
- Make sure to plate a negative control next time.
Gene synthesis assignment: Patrick, Sherine
SOEing PCR assignment: Susan, John, me
Basic PCR assignment: Elicia, Joey, Tom
SOEing parts: ig302, ig306, ig310, ig314, ig318, ig322
PCR #1:
- 12 tubes (12 rxns), 24 oligos
- 14 x cocktail: 336 uL ddH20, 46 uL 10x Expand Buffer 2, 46 uL dNTPs, 7 uL Expand Polymerase 1
- To each tube add 31 uL of cocktail, .5 uL template DNA, 1 uL of each oligo
- tubes labeled 302A, 302B, etc. corresponding to two reactions per part (ex. 302A: add oligos 302/301; 302B: add oligos 300/303)
- Mix tubes with pippet
- Set up PCR: all tubes except 314B ran in program 2K55, 314B ran in program 4K55 ( >2kb )
Wednesday, June 24
Redo 306 yet again...
- Try new genomic DNA: Mgll55
- Oligos:
| Part | Oligos | Location | Length of Part |
| 306A | 306/305 | G1/F1 | 464bp |
| 306B | 304/307 | E1/H1 | 452bp |
Mixture cocktail for setting up SOEing PCR1 (x3)
- 72 uL ddH20
- 9.9 uL Exp "2"
- 9.9 uL dNTPs
- 1.5 uL Exp pol "1"
Try adding 1 uL DMSO next time if it doesn't work again
Place in block A at 10:46 am (2K55 program)
EcoR1/BamH1 Digestion of 302, 310, 318:
- 4x cocktail mixture
- 2 uL into each PCR tube labeled 302, 310 318
- incubate 37 degrees for 1 hr. (start: 11:05 am end: 12:05 pm)
Redo 306 yet again...found a new template DNA that should work!
- Same procedure as earlier today
- put in 2K55 at 11:17 am
Check 24 block plate from yesterday:
- Miniprepped colonies from 57A, 113B and 129B
Ligation: 302, 310, 318 with vector pBca9523
- incubate 30 min on bench top 12: 19 (end at 12:50 pm)
- MESSED UP, did not add from digestion tube....redo and start incubating at 12:28 pm (end 1:00 pm)
Transformation by heat shock: 302, 310, 318
- LB (rescue) from upstairs
- Plates (Spec) from upstairs
- Spreading method --> beads?
- Put in incubator at 1:10 pm, take out at 1:50 pm
Plating 302, 310, 318 (with Jenn and Susan)
- 100 uL ligation
- New LB
- New plates
- Spread with loop on downstairs bench with benchtop flame
Gel purify 306 PCR
- Lanes: 1. ladder, 2. 306A, 3. 306B
306: zymo gel purify, elute 50 uL, 2nd PCR: oligos 306/307 placement G1/H1 on plate
For samples 314 and 322: after PCR 2 and analytical gel Okay, did a regular zymo clean up (add 180 uL ADB buffer and eluted with 33 uL ddH2O
- Then run a digestion
- run agarose gel; melt with 600 uL ADB buffer (55 degrees)
Thursday, June 25
Did some general tidying up.
Retransform with TG1 cells.
Pick colonies from yesterday.
- Before retransforming:
- inserts gel purified after digest --> ~6 uL
- vector --> digest --> gel purify --> ~30 ul
- ligate --> 10 ul
- transformation changes: TG1 cells, cleaner benchtop, bead spreading/L shaped, cleaner EtOH
Redo ligation and transformation: 302, 306, 314, 322
Vector: pBca9523-1144#5
- Re-digestion: 30 ul
- Run gel: 10 ul, 2 ul loading dye
- 6pm transform
- 314 tube gone?
Friday, June 26
Saturday, June 27
Sunday, June 28
Monday, June 29
Tuesday, June 30
July
Wednesday, July 1
24 well block with samples 27, 57, 79, 113, 121, 137, 145, 213, 239, 261, 302, 314 left overnight to grow from yesterday, grew very well...lots of cells!
- Miniprep 24 samples (each part in duplicate)
Monday, July 6
Set up colony PCR reactions for 213 and 239 for colonies that Susan plated last night.
- We ran out of forward and reverse primers.
- Made new 10x dilutions of primers.
When we get back from bowling: run gel from col pcr that Mahdvi set up last night and then miniprep and send out for sequencing; possibly set up a PCA from scratch for 213 and 239.
Tuesday, July 7
Set up PCA reactions from scratch for 213 and 239. Assembly step used oligo mixes from freezer.
Work on TECAN assay:
- Susan induced 18 samples in 500 uL of LB for the TECAN assay this afternoon.
- We decided not to run in triplicate.
- For each sample, various concentrations of streptavidin will be used (0, .25, .5 and 1 uL concentrations)
- The positive control to be used in pBca- 9494, negative control will be uninduced 9494.
- The concentration of the cells will be determined by taking OD measurements of the cells in the TECAN.
Talk to Susan and Patrick about starting to work on a model for the project. Should bring up at the next meeting.
At lunch, John Wang told us that he discovered that the primers for 213 and 239 were switched. 213F and 239R were switched. John, ran gels to confirm this.
Susan zymoed John's PCA products and set up a digestion.
I set up the second round of PCR amplification for the 213 and 239 reactions started this morning. All four primers were added to each tube (both 213 and 239 forward and reverse primers) just in case tube switching did occur and it was not properly fixed.
- We should get to plating John's 213 and 239 products and through the TECAN experiment this afternoon.
Ran analytical gel on 213 and 239 PCA products and they were great! So, we have back-ups just in case John's samples don't end up working out.
TECAN:
- We set up the plates with 100 uL of cell culture for each condition being tested.
- Streptavidin dilutions were performed by adding 35 uL of strep to 315 uL of 1X PBS.
- Susan and I decided to place the samples in the fridge until later (8:30 pm) when we could come back and finish the assay.
- Added 2.5, 5, and 10 uL of strep diluted 10X to appropriate well in 96 well block.
- Incubated for 30 minutes.
- The following cells did grow: M1020, M10211, M10212, M10213, M10214, M10215, M10216, M10217, M10218, M10219, M10221, and M10224; as well as 9494 as a positive control (and uninduced negative control).
- The plates were balanced before centrifuging and then the supernatant was removed and the cells were resuspended in 100 uL of 1x PBS. This was performed twice. After the second resuspension, the liquid was transferred to the black-sided, clear flat bottomed plate for TECAN measurements.
- Susan and I couldn't find the file for taking measurements, so we decided to cover the plate in foil and place it in the fridge overnight.
Wednesday, July 8
Plans for today: work on TECAN assay with Lane, grow up and miniprep John's 213 and 239 plasmids that were plated yesterday and send them out for sequencing, work on powerpoint slides for group meeting tomorrow afternoon, research functional assays for TirM and caspase.
Susan came in earlier this morning (at 7 am) to pick colonies from 213 and 239 plates. They should be ready for miniprepping at approximately 5 pm.
Took fluorescent readings from the flat-bottom plate in the TECAN, using Terry's measurement parameters file: RPE_new. The trend was not quite as expected. We suspect there are many false positives due to inconsistencies in pipetting and residual streptavidin remaining in the wells. We cannot confirm this because we didn't add strep to our negative control.
Spoke with Lane about the TECAN results and he wants us to run the assay again, and compare the flat bottom results to the v-bottom readings.
TECAN assay work:
- First run through: set up two plates (one v-bottom, one flat-bottom) with duplicates of each sample.
- Decided to use same concentration of strep in all wells.
- Took OD measurements, and saw a decrease in absorbance in v-bottom plate (as expected).
- Then pelleted the cells...saw strong pellets in the controls, but tiny pellets in the parts. No visible pellets in the flat-bottom plate
- Talk to Terry, and decide to use flat-bottom plate at the very end, to do comparison: after taking fluorescent measurements in v-bottom plate, transfer liquid to flat-bottom plate and retake measurements.
- Made strep dilution: 1.9 ml of 1x PBS plus 19 uL of concentrated strep
- Incubate and then follow protocol as described online.
- Note: Terry showed us how to use the robot to add PBS in the wash steps: go to run -> prime, then run-> dispense-> 03 -> enter -> start.
- The Image taken for the first strep assay run through after 1 wash with PBS:
The cells in A1 and A3 showed a higher degree of fluorescence, which was later supported by the TECAN data taken both in the v-bottom plate as well as the flat-bottom plate.
- Second run through: set up one v-bottom plate with triplicates of each sample: A1-A4, 9494 and 1363 (all induced).
- Higher concentration of cells...we let them grow up for approximately 8 hours instead of 5.
- Used the same concentration and dilution in PBS as in the previous experiment.
- Took initial OD600 measurements, then pelleted the cells. Saw much larger pellets than in the previous run through. Might try growing up overnight to saturate cells.
- Followed Terry's protocol after that.
Thursday, July 9
Prepared powerpoint slides analyzing the TECAN/imaging strep assay results from yesterday. Once the data was normalized to OD600 measurements, the A1 fluorescence, which had a much smaller pellet, was comparable to that of A3. The data for the v-bottom was proportional to that of the flat-bottom plate, so we will probably be using the v-bottom plate to avoid fluid transfer step to the flat-bottom plate.
Meeting at 2pm with iGEM team for weekly update:
- I will be working with Jenn on the robot and composite parts.
- The strep assay: we have decided to stay with the v-bottom plates as they provide similar data as the flat-bottom plates and enable qualitative pictures as well as quantitative TECAN data to be collected.
Completed a restriction map with Jenn on 8 parts.
The gel looked pretty good. The smaller bands are difficult to see, but since the DNA was diluted more than usual, the results point to successful miniprepping. We will be using these strips for the rest of the parts as we continue with composite parts.
Friday, July 10
Spoke with John W about the signal transduction systems. I will be working on some literature searching in parallel to the robot work. There are three systems that we are looking at for the signal transduction: Tev-N and Tev-C, Nub and Cub, and caspase. They will incorporate leucine zippers and the Tox-R system for signal transduction. There are a few problems associated with the systems, such as binding of the B part of the ABA trimer system before it reaches the outermembrane of the cell. To reduce this background noise, we hope to introduce a signal amplification system with another enzyme that acts on B, cleaving it from the membrane.
Met up with Jenn in the lab and she was working on Miniprepping the samples that were transformed last night into the in-vivo Gateway cells. We hope to perform a restriction map on 8 of these samples that are miniprepped and then, if successful, transform righty and lefty cells today.
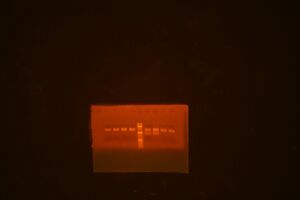
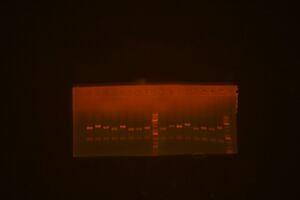
Following Jenn's minipreps, I set up a restriction map digestion for DNA in the following miniprep wells: A1, A3, A5, A7, A9, A11, B1, B3. The gel image is found below:
- The gel lanes: 1) A1, 2) A3, 3) A5, 4) A7, 5) ladder, 6) A9, 7) A11, 8) B1, 9) B3
Sunday, July 12
Learned how to make competent cells with Mahdvi:
- Pour 1:100 cells: LB in the large flasks (one for righty, one for lefty cells)
- Incubate in the 37 degree room upstairs for 2-2.5 hours
- Take OD reading (both were around .77); want a reading within .5 and .8 range
- Pour culture into smaller bottle to pellet cells in centrifuge (A-10, 6500 rpm for 5 minutes at 4 degrees C)
- Pour supernatant LB back into large flasks to bleach clean later.
- Pour 25 mL of TSS into each of the smaller bottles to resuspend and put on ice.
- Label tubes for aliquots
- Pour 1.5 mL into each 2 mL tube in the 4 degrees C room.
Picking colonies and spotting on spec plates:
- Examined A/B strip plate:
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
| A | N | 1 | N | Y | N | 7 | 2 | N | Y | Y | 6 | 1 |
| B | Y | Y | 5 | N | Y | Y | Y | Y | Y | Y | Y | Y |
- C/D strip plate:
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
| A | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| B | Y | Y | Y | Y | Y | Y | Y | N |
Setup 3 96 well blocks and 3 spec plates with A 1-12 corresponding to A 1-12 on the strip plates as well as with B, C, D.
The following were blanks (no colonies picked to grow up):
- Block 1: A1, 3, 5, 8; B4
- Block 2: A1, 2, 3, 5, 8, 12; B4
- Block 3: A1, 2, 3, 5, 8, 12; B4; E3
Made an error in the 3rd replicate plate: accidentally placed the toothpick for D3 in D4, leaving D3 empty. The colonies were shifted over by one in the 3rd plate.
Monday, July 13
List of things to do today:
- Pour LB agar plates: regular spec, large spec, spec AK, spec AC, spec CK
- Need to re-transform into gateway cells: liquid culture -> plate set; plated set; and plated -> liquid culture set.
Possibly restriction map those parts that did not transform well into the lefty and right cells. (Didn't end up doing this)
Examine spotted plates from Sunday. (I punctured the agar too deeply, making it difficult to distinguish cells that grew) The following were the locations on the spec plates that showed growth:
- Plate 1: B1, B2, B6; C6; D7
- Plate 2: A6, A7; B6; C4, C10; D4, D7
- Plate 3: A4, A6, A11; B6, C10; D5 (which is D6 on the block)
- B6 was bad for all three; A6, C10, D7 were bad for two.
Failed parts from gateway to retransform (located on basic entry plate):
- C2(location on basic entry plate) <cel3a!> A1(location on LB agar strip plate) in AK cells
- C3 <cel5b1> A2 in AK cells
- C4 A3 in AK cells
- C6 <TirM> A5 in AK
- B1 <mgfp> A7 in AK
- A8 <needle scFv> A8 in AK
- D2 <cbk> A12 in AK
- D5 <beta helix> B3 in AK
- D6 <glp LVA> B4 in AK
- D8 D8 in KA
Took 4 pir AK and 2 pir KA cell aliquots from -80 C freezer. Thawed on ice and then added 10 ul of KCM and no water to each 90 ul aliquot (at first added 30 ul KCM and 50 ul ddH2O, had to get new set of cells).
Spot on large spec plates using pin tool:
- There were visible cells immediately in the following positions following spotting:
- Plate 1: C5, C11, C12, D4
- Plate 2: C11?
- Plate 3: C1?, D1? . . . not quite as many cells in these (fainter dot)
Tuesday, July 14
Plates from yesterday look good (all except A12)
Examine spot checking plates for co-transformation:
- Plate 1: B1, B2, B6, C6, D7
- Plate 2: A6, A7, B6, C4, C10, D4
- Plate 3: A11, A7, A6, A4, B6, C9
In 24 well block: A12 did not grow.
Plans for today: miniprep, restriction map, transform into pir Righty cells from 24 well block:
| A1 | A2 | A3 | A5 | A6 | A7 |
| A8 | A12 | B3 | B4 | B6 | D8-1 |
| D8-2 |
Did not need to re-miniprep: A6, A7, A12, B3 (they worked in the first set)
Miniprepped remaining 9 parts: A1, A2, A3, A5, A8, B4, D8-1, D8-2
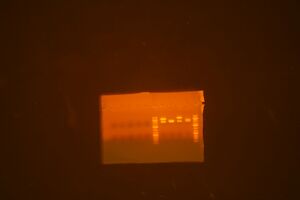
Performed a restriction map digest (Eco/Bam) on the 9 parts:
- A1 <cel3A> AK righty 2541 bp
- A2 <cel5b> AK righty 1599 bp
- A3 <cel9a> AK righty 2796 bp
- A5 <TirM> AK righty 327 bp
- A8 <needle scFv> AK righty 837 bp
- B4 <GFP-LVA> AK righty 756
- B6 <GS5-IILK> AK righty 147 bp
- D8 <dbl term> KA righty 135 bp
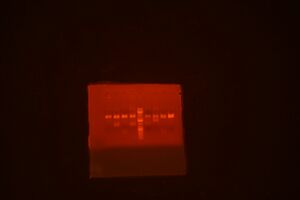
Gel pic:
Lanes from left: 1) A1, 2) A2, 3) A3, 4) A5, 5) A8, 6) ladder, 7) B4, 8) B6, 9) D8-1, 10) D8-2
- Gel looks pretty good. The smaller parts are difficult to see, but that is expected.
Perform a transformation of miniprepped plasmids into pir Righty cells:
- Dilute miniprep: add 1 ul of DNA to 19 uL of ddH2O
- Add 30 uL of KCM and 50 uL of ddH2O to the cells.
- in PCR tubes add 30 uL of cell mixture and then 1 uL of plasmid dilution
- rescue in 100 ul of 2YT for 20-25 minutes
- plate 100 uL on AK plates
Stored minipreps in Jenn's brown freezer box. (Will add to stock plates tomorrow depending on plating outcome)
Wednesday, July 15
Check plates for A1, A2, A3, A5, A8, B4, B6, D8-1, D8-2 transfers:
- All plates had lots of colonies! :)
Assembly set on strips that Jenn plated:
- three parts did not work; Susan is catching them up by re-assemblying them
Pick colonies from transfer plates of 9 parts:
- Add 50 uL of 2YT with AK antibiotic to three rows (A-C) of a 96 well block.
- Use toothpicks to pick 4 replicates of each transfer into the 96 well block as follows:
| A1 | A1 | A1 | A1 | A2 | A2 | A2 | A2 | A3 | A3 | A3 | A3 |
| A5 | A5 | A5 | A5 | A8 | A8 | A8 | A8 | B4 | B4 | B4 | B4 |
| B6 | B6 | B6 | B6 | D8-1 | D8-1 | D8-1 | D8-1 | D8-2 | D8-2 | D8-2 | D8-2 |
Start growing up in shaker a little before 11:30 am. Should spot around 4:30 pm.
Realized that 50 ul is not enough liquid in the 96 well block for the pin tool to reach the bottom of the well. Added 1 mL of 2YT with AK to each well and placed in shaker at 2:45 pm. Need to let the cells grow for about 2-3 hours before spotting on spec plates.
Thursday, July 16
Examined spec plates spotted for co-transformation:
- Growth seen in C4, C6, C10, C12 (<GS5-IILK> and <dbl-term> parts in pir righty cells)
Set up a restriction map digestion for parts: Aa AII ATD(E5), eha B(E7), virG ATD(H7), dbl term(D9):
- These colonies showed co-transformation
- Digested E5, E7, H7 with BamHI/XhoI
- Digested D7 with BglII/XhoI
- Part info:
- E5: KC (2072/6820)
- E7: KC (2072/7807)
- H7: KC (2072/8008)
- D9: (2699/6162)
Pick colonies from C/D iGEM09 Basic Assemble RL: D6 <pBad.rbs.prepro.streptag>
- Picked 5 colonies into 1 ml of 2YT + AC
- We think that there may have been a mutation in the ccd gene in the colony picked previously from this plate
Some plans for today: re-gateway and pick colonies for:
- <pBad.rbs.prepro.streptag> (only re-gateway)
- <VtaII AtD> (re-gateway and re-transform)
- <EhaB> (re-gateway and re-transform)
- <virG> (re-gateway and re-transform)
Also, want to miniprep and restriction map pir Righty basic parts that I transformed and grew up yesterday and the assembled parts that did not show co-transformation on Jenn's plates.
Spoke with John W about prioritizing some parts and we decided to restriction map them with BamHI/XhoI:
- D3 <cub> (6300/2072)
- E3 <nub> (6309/2072)
- F3 <casp> (6996/2487)
- G3 <tevN> (6522/2487)
- H3 <tevC> (6540/2487)
- A5 <prepro> (7620/2487)

For assembly sets performed miniprep with strips:
1) Spin down 5,500 rpm for 5 minutes (balance before placing in centrifuge)
2) Flick out liquid
3) 250uL A1; vortex (next time try to use P1 to more easily see color change)
4) 250ul A2; vortex
5) 350uL A3; vortex (for a while)
6) spin down 5,500, 5 min
7) set up strips w/pink silicon base
8) transfer supernatant (being very careful not to pick up the cell junk at the bottom of the tube; if accidentally pick some up, re-pellet and try again)
9) Spin 2500, 2 minutes ... continue with protocol as usual (on pcr plate will use every other column due pink base constraints)
Friday, July 17
Miniprepped and Restriction mapped:
- <azo> <term>
- <Hag> <term>
- <upaG> <term>
- <AIDA> <term>
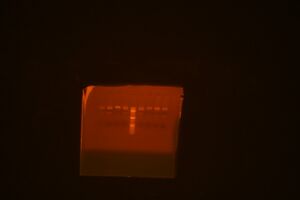
Restriction map digest with both BamHI/XhoI and BglII/XhoI (BamHI digests are the first four on the left, BglII digests are the second set of four on the right after the ladder)
Monday, July 20
Over the weekend the team accumulated some more data and there appear to be problems consistently linked to the KC lefty parts. We also think that there maybe some problems with the miniprep strips not giving clean enough DNA. Today we hope to figure out what the source of error is.
Split up duties:
1) PCR composite and basic parts of the <disp><term> set of composite parts.
2) Conduct miniprep tests on pBjb1601KC parts that were transformed into pir lefty as well as pBdr052 plasmids that mapped well in righty cells and were transformed into pir lefty cells. Use BamHI/XhoI to restriction map
3) Miniprep composites <internal passengers><term> in righty cells and then digest with BamHI/XhoI.
4) Run a test on the enzymes themselves.
- I was in charge of #2
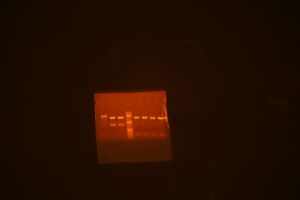
I miniprepped and restriction mapped the pBjb and the pBdr052 parts in the pir lefty plasmids:

Lanes from the left: 1) A2, 2) B4, 3) C1, 4) D1, 5) ladder, 6) pBjb21-ca, 7) pBjb2127-ca, 8) pBjb217-kc, 9) pBjb2128-ca
- B4 did not grow well; the cell pellet was quite small. (probably why that lane is empty)
Miniprep on strips to compare and try to improve on the protocol that has resulted in poor digests.
- Used aluminum holder for columns
- washed with A4 twice
- Used Row D1-5 from plate: composite parts iGEM C09ig0019-C09ig0023 <4 reps each> 7/18 as well as three parts from earlier today: A2, C1, D1
- For wash steps (AW, and A4 twice) centrifuged at 2500 for 2 minutes
- To dry the column, centrifuged at 2500 for 5 minutes
- Eluted with 100 uL of ddH2O into first column of a PCR plate: A1-E1 correspond to D1-5 in the composite parts plate; F1 is A2, G1 is C1, and H1 is D1 from lefty cells.
Ran a restriction digest for mapping. A1-E1 were mapped with BglII/XhoI, and F1-H1 were mapped with BamHI/XhoI:
Lanes from left to right: 1) A1, 2) B1, 3) C1, 4) D1, 5) ladder, 6) E1, 7) F1, 8) G1, 9) H1
Tuesday, July 21
Today I picked colonies from the iGEM 09 Assembly that Jenn set up last night. There were 4 plates of strips (A-G, 1-12 for each row) and B6 and B7 were plated on separate dishes because they were plated on the wrong antibiotic in the strips.
Wednesday, July 22
Colony PCR from yesterday was not very clear (Patrick diluted the dNTPs to 1mM instead of 2mM and the 4k55 program was run instead of the colony PCR program), so we decided to re-run the colony PCRs.
I ran the gel for the 3 replicate sets of 6 assembly products that we chose from yesterday:
Gel analysis:
- A4 (3474 bp) clone 1: good; clone 2: good; clone 3: cotransformation
- A12 (2214 bp) clone 1: cotransformed; clone2: cotransformed; clone3: no bands
- B1 (3999 bp) clone1-3: good
- B4 (2214 bp) clone1: cotransformed; clone2-3: good
- C4 (1866 bp) clone1-3: good, running a little big
- E8 (2544 bp) clone1: good, a little big; clone2: good, not as bright; clone3: cotransformed
Thursday, July 23
Today is a robot assembly day. I will perform assembly on the mussel foot / scFv parts with Jenn's assistance. This should be the last round of assembly for these parts as well as the cellulase parts before being able to run some assays. This morning Jenn had started assembly of the cellulase parts and guided me through the steps of robot assembly. Some notes from this are:
- Files necessary for robot assembly are found on the desktop in a Biomek everything folder. Within this folder is a temp files folder that has a 2AB guide by Jenn file that contains a brief outline of the assembly protocol. The red steps in the protocol are robot steps.
- For the dilution step using miniprep strips: want a 2x dilution of plasmid in 1x Neb2. The Neb2 is in the fridge and has a 10x concentration, which must be diluted to a final concentration of 1x in the Bam/Bgl cocktail (example: for a final volume of 20 ul, add 2 ul Neb2, 8 ul of water, and 10 ul of plasmid).
- For the digestion reaction set up: calculate the total number of reactions, make Bam/Bgl cocktail in cryo tube (make a little more than needed) and place in A6 in P4, place a fresh 96 well plate in the destination plate location, place 96 well dilution plate in source plate location.
- To run the program: go to open file in the biomek program (making sure that the csv file to be used is closed before running it), go to transfer from file and choose the correct csv file, go to instrument set up and make sure the pipette tips diagram is accurately filled.
- Incubate (cover with foil) in thermocycler: Run -> JCA -> 123
- Continue with protocol in the 2AB guide by Jenn file
- For the rescue step, add 100-150 ul of 2YT to a 96 well block and then use a multichannel pipette to transfer the transformed cells from the PCR plate to the 96 well block. Incubate in the shaker for 45 minutes to 1 hour.
- When performing assembly, can stop after the digestion step by covering with foil and placing plate in freezer, however, after ligation, must continue to plating.
- Save the 2ab-LR files for records
- When making the ligation cocktail, use 10 mM ATP found in green box in door shelf of the freezer. Make sure to return to freezer after using it.
Friday, July 24
Got sequencing results back on a few composite assembly parts and all were correct except the <mgfp> part, which sequenced as super oxide dismutase (SOD is located in C1, and mgfp is in B1 so I might have accidentally sent the wrong plasmid in for sequencing). We are sending in both B1 and C1 in for sequencing today. We have also decided to sequence the RL products and gateway products of mgfp to trace the error step.
Checked the plates for the final round of assembly for the cellulase, mussel foot and scfv parts and there were colonies in every strip. We picked colonies into 1 ml LB + CA and will grow them up for spot checking.
We also plan to grow up some competent cells of EC100D pir + cell line used for yesterday's assembly transformation.
- Didn't have time to make competent cells, will make them tomorrow.
Saturday, July 25
Checked on the spot check plates and very few clones showed cotransformation. This means, we can prepare these cells for assaying tomorrow.
We will start the final round of assembly for the parts that are catching up on the robot.
The sequencing results showed that both B1 and C1 in the composite assembly parts plate were both SOD. Also the wrong part appears to be in the RL cell, but gateway appears to be all right. We will transform the gateway part into the pir righty cell.
Make competent cells of EC100D pir +.
Sunday, July 26
I came in to lab to pick colonies from the internal passengers assembly set that we ran yesterday.
- There were a few strips that did not have any colonies, and others only had a couple.
- I picked three clones when possible making three plates. A12, C2, and D4 were empty wells for all three clone plates. For clone plate 3, G2 was also empty.
I also checked the competent cells from yesterday and there appears to be contamination on the spec plate. The rest of the plates looked fine. So I re-picked a colony from the streaked plate of EC100D pir plus to grow up in 5 ml of LB to re-try making another batch of competent cells. <mgfp>
I re-transformed <mgfp> into pir righty (located in F1 in the Basic Assemble plate) and plated on KC and directly into LB+KC.
I also miniprepped the prepro part that grew up yesterday.
Monday, July 27
Made more competent cells from the EC100D pir + culture that was grown up yesterday and overnight. Placed in the 37 degree room on the shaker at 10:40 am. Should grow for approximately 2.5 hours. The OD measurement at 1:25pm was 0.648, which is within the acceptable range.
Miniprepped mgfp in pir righty (pBdr052AK-B09ig239) that was grown up yesterday by Jenn (she had told me the wrong antibiotic combination, so she re-transformed into AK last night).
Perform layered assembly on mgfp with pBad (pBdr052CA-1363) located in C4 of the RL1 plate in the minifreezer. Added the following to a PCR tube:
- 1 ul lefty
- 1 ul righty
- 1 ul NEB2+ATP (tube with red stripe on the top)
- .3 ul of XhoI, BglII, and BamHI
- 6.1 ul of ddH2O
Digest for 1 hr, then heat kill for 20 minutes at 65 (this program is called 123 on the black thermocycler).
Add .3 ul of T4 DNA ligase and then incubate on the bench-top for 30 min (start 1:19pm, end at 1:49 pm).
Transform into pir lefty (yellow tube with red stripe on the top) and then plate on CK antibiotics.
Perform colony PCR on six parts from the internal passenger assembly set:
- <tevc> in A1
- <tevN> in B7
- <caspace> in C9
- <nub> in D11
- <cub> in F4
- no pass in G5

The order was: clone1, ladder, clone2, ladder, clone3, ladder. The parts were in the order listed above from left to right for each clone.
Following the 4pm meeting, I started a EcoRI/BamHI transfer protocol for our positive control, 9494 into pBca9523. The layered assembly protocol for EcoBam transfer was to add the following to a PCR tube:
- 7.5 ul ddH2O
- 1 ul of NEB2
- .5 ul EcoRI, BamHI, and plasmid
Place in thermocycler using the ecobam program that incubates at 37 for 1 hr and then heat kills at 80 for 20 min.
Add .5 ul of digested vector and .5 T4 DNA ligase; then transform and plate as usual. (We asked Lane to take over after the ligation step.)
Tuesday, July 28
Today we need to organize and figure out where to go next.
After checking the mgfp plate from yesterday (not sure if it was a lawn or if nothing grew), we decided to re-assemble <mgfp> with the pBad.prepro part. Follow the same protocol as yesterday with the layered assembly. I used 1 ul of each plasmid and all three restriction enzymes (Bgl, Bam, Xho). I transformed the 10.5 ul into 70 ul of pir lefty cells by heat shocking and then rescued in 100 ul of 2yt for 45 minutes.
Checked the EC100D pir + plates from yesterday and spec once again had some small colonies growing. There were 2-3 colonies also on Cam. I picked a few colonies from the streaked plate from 7/23 into 1 ml of LB. These will be plated on each antibiotic plate to see if they are contaminated.
I tried restriction mapping some parts again, to see what is going on with the parts that were sequenced. These are the parts:
- F09ig0011 clone 1, sequenced as the parent vector
- F09ig0024 clone 1, sequenced perfect
- F09ig0205 clone 1, sequencing as the parent vector
- F09ig0141 clone 1, sequenced perfect
Also miniprepped F09ig0011 clone 3 and F09ig0205 clone 2 to restriction map.
Wednesday, July 29
I came into lab at around 4pm after completing some training in the machine shop and was filled in on the discoveries made throughout the day:
- The pbad promoter in the pBdr052 plasmid was activating phagemids that were killing the cells, so we are going to switch over to pBca9495. This takes us back one step in the assembly tree.
- Also, it turned out that the EC100D pir + cells were spec resistant from the stock that was streaked onto an agar plate.
I miniprepped pBca9523-Bca9494 and restriction mapped with Eco/Bam.

Thursday, July 30
Checked streaked plates of righty and lefty cells and they looked good. I picked a colony from each plate to grow in 5 ml of LB. They should be ready in about 12 hours (10:30 pm), will probably grow them up tomorrow.
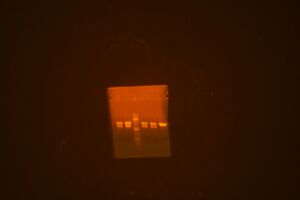
Run gel of 9495 vector backbone parts: M10303 in pBca9495AC (3048 bp), M10301 in pBca 9495AC (3048), M10332 in pBca9495KA (3180)
The lane order goes: 003, 322, 322, ladder, 301, 301, 303, 303
Friday, July 31
Miniprepped A4, B4, C4 from pBca 9495 plate (placed in A2, B2, C2)
Started making competent cells of the righty and lefty strains.
- Make TSS by preparing and autoclaving the following seperately:
- 85 ml LB
- 10 g PEG-3350 in 5 ml H2O
- 5 ml DMSO
- 2 ml 1M MgCl2
- Allow to cool and then combine and fridge
- OD measurements of cells at 2:25 pm:
- Righty: 0.6221
- Lefty: 0.581
- Susan, Patrick and Gabe helped in 4 deg room to make cell aliquots
- Plated 50 uL on each antibiotic plate and transformed with amp resistant plasmid with RFP as a test for contamination.
Set up EcoRI/XhoI Digest for restriction mapping A1-H1 and A3-H3 (B1, F3 are empty)
August
Saturday, August 1
I came in to miniprep the EC100D pir + plasmids using the 8 strip miniprep columns. The set-up for the miniprepped PCR plate is located in the iGEM09 Parts google docs page in the pBca9495 Beginnings sheet under 96 well miniprep plate position.
I also set up the EcoRI/BamHI transfer reactions for the righty parts that I miniprepped.
Monday, August 3
Miniprepped plate from righty transformed pBca9495KC displayer parts.
Ran a restriction map with EcoRI/XhoI as well as BseRI/XhoI(see Jenn's notebook):
The lane order goes: C09ig0027, 35, 24, 25, 26, 28, 29, 30, ladder, 31, 32, 33, 34, 36, 37, 39, 40, ladder.
Expected sizes are
- pBca9495 KC C09ig0027: 2219/1142
- pBca9495 KC C09ig0035: 3206/1142
- pBca9495 KC C09ig0024: 2810/1142
- pBca9495 KC C09ig0025: 2660/1142
- pBca9495 KC C09ig0026: 2804/1142
- pBca9495 KC C09ig0028: 2219/1142
- pBca9495 KC C09ig0029: 2222/1142
- pBca9495 KC C09ig0030: 2219/1142
- pBca9495 KC C09ig0031: 2246/1142
- pBca9495 KC C09ig0032: 2840/1142
- pBca9495 KC C09ig0033: 2840/1142
- pBca9495 KC C09ig0034: 3329/1142
- pBca9495 KC C09ig0036: 2453/1142
- pBca9495 KC C09ig0037: 3551/1142
- pBca9495 KC C09ig0039: 2849/1142
- pBca9495 KC C09ig0040: 3401/1142
Parts C09ig0027, 30, and 37 look incorrect. I miniprepped the second clone of each part and then ran a restriction map using the following volumes per reaction: 5 ul of DNA, 12 ul ddH2O, 2 ul of NEB2 Buffer, 0.5 ul XhoI, 0.5 ul EcoRI or BseRI (total volume of 20 ul). Digest at 37 for 2 hours.
I also picked four colonies of righty and lefty streaked plates to grow up in LB for competent cells and to test for contamination on each of the antibiotic plates. I plated 10 ul of the culture and let grow overnight.
Tuesday, August 4
Checked righty and lefty plates and there appears to be no contamination. I will make competent cells tomorrow with autoclaved LB in the large flasks.
Jenn ran the restriction map gel that I set up last night. Clone 2 of part C09ig0030 still looks funny. Sherine miniprepped this and let digest for 30-60 minutes. We want this part to be ready before we move forward with 2AB reactions on the robot.
I autoclaved (45 minutes liquid cycle) a few bottles of 2YT to add antibiotics to as well as a couple bottles of LB agar in 500 ml jars to pour strips later today.
Set up 2AB reactions for cellulase and scfv/leucine zipper parts. There are 68 cellulase parts and 87 scfv/leucine parts. Go through to plating
Wednesday, August 5
The cellulase and scfv/leucine zipper strip plates looked good. There were plenty of colonies for each part. Parts with displayers CPG_L6 and yuaQ consistently had fewer colonies.
Autoclaved 24 and 96 well blocks for picking colonies this afternoon.
I picked two clones of each cellulase part while Susan and Sherine miniprepped eco/bam transfers of internal passengers. Jenn then picked two clones for each scfv/leucine zipper part into two 96 well blocks.
Should be able to spot check on large AKC plates later today.
Thursday, August 6
All <yuaQ AtD> and <CPG_L2> parts showed cotransformation on the spot check plates from last night. I ran a restriction map digest on the plasmids from 8/3 to determine if the parts going into assembly look funny.
I picked a colony from the DH10B cells that I plated for making competent cells. Should be able to make some competent cells tomorrow.
I miniprepped samples for sequencing from the cellulase and scFv/leucine zippers final parts clone one blocks:
Cellulase:
- A1: F09ig0139 {Pbad.rbs.prepro.StrepTag}{<cel3A!}{<azo1653 AtD>}(dblterm)
- B6: F09ig0157 {Pbad.rbs.prepro.StrepTag}{<cel5B!}{<OprF AtD>}(dblterm)
- C11: F09ig0175 {Pbad.rbs.prepro.StrepTag}{<cel9A!}{<cl02365 AtD>}(dblterm)
- E4: F09ig0193 {Pbad.rbs.prepro.StrepTag}{<cel6A!}{<VtaA11 AtD>}(dblterm)
scFv/Leucine zippers:
- A5: F09ig0005 {Pbad.rbs.prepro.StrepTag}{<type IIIs Needle Complex scFV>}{<Hag AtD>}(dblterm)
- B10: F09ig0023 {Pbad.rbs.prepro.StrepTag}{<gliadin binding scFv>}{<Pcryo_1225 AtD>}(dblterm)
- D3: F09ig0041 {Pbad.rbs.prepro.StrepTag}{<KILR>}{<Hia AtD>}(dblterm)
- E8: F09ig0059 {Pbad.rbs.prepro.StrepTag}{<EILD>}{<upaG_short>}(dblterm)
- G2: F09ig0078 {Pbad.rbs.prepro.StrepTag}{<GS5-IILK>}{<CPG_L6>}(dblterm)
Ran eco/bam digest of minipreps (20 ul protocol)
Lanes are as follows: F09ig0005, 23, 41, 59, 78, ladder, 139, 157, 175, 193. I don't know why there is no DNA in the first five lanes (this was the second set of minipreps that I performed). Maybe there is something wrong with a reagent on our side of the bench. I decided to quickly miniprep another set of the same parts from the 96 well block of clone 2 as Gabe offered to drop off these samples to the sequencing facility (this set in the sequencing log are: iG037-046). The cellulase parts have sequencing numbers iG029-iG036. iG019-iG028 were canceled, as the gels showed there was no DNA in the miniprep products.
Friday, August 7
Run and eco/bam digest of parts F09ig 5, 23, 213, 231, and 250. (213, 231, and 250 were mislabeled on the final parts google doc spread sheet as 41, 59, and 78; we relabeled everything to fix this)
Lanes: 5-1, 23-1, 213-1, 231-1, 250-1, ladder, 5-2, 23-2, 213-2, 231-2, 250-2.
Once again, there is no DNA in the first lanes.
Saturday, August 8
Need to:
- Pour CA strips
- complete 2AB assembly from ligation to plating (93 rxns total)
- use DH10B cells for transformation (if there are not enough DH10B, use MC1061 or TG1); if there are 93 rxns, need 19 tubes
- transform with 50 ul cells and rescue in 100-125 ul of 2YT for 45 minutes
Monday, August 10
Pick colonies and streak those that were lawns. The parts were internal passengers, intermediates, and mgfp parts. The parts that were lawns and were streaked on CA plates were: B2-B12, C1, C2, E5-E12, F1-F7, F9-F12, G1, G3, G10-12, and H1-3.
Tuesday, August 11
Spot check plates showed little cotransformation for mgfp parts. Some mgfp parts grown in 2YT+AC need to be miniprepped for restriction mapping and sequencing. F6-G9 is set one, G10-H12 is set two from mgfp parts.
Realized that there was a mistake made on Saturday. I thought the 2AB assembly was only of final parts, and when I asked what cells to use to transform, I was told to use DH10B, so I continued to assume that they were all final parts when in actuality there was a set of intermediates. We will have to redo the 2AB reactions for the internal passengers and displayer intermediates from Saturday.
Minipreps:
- F9 clone 1 set 1: F09ig0261 {Pbad.rbs.prepro.StrepTag}{<mgfp-5>}{<VtaA11 AtD>}(dblterm)
- H1 clone 1 set 2: F09ig0261 {Pbad.rbs.prepro.StrepTag}{<mgfp-5>}{<VtaA11 AtD>}(dblterm)
- F11 clone 1: F09ig0263 {Pbad.rbs.prepro.StrepTag}{<mgfp-5>}{<Hag AtD>}(dblterm)
- G1 clone 2: F09ig0265 {Pbad.rbs.prepro.StrepTag}{<mgfp-5>}{<upaG_short>}(dblterm)
- G5 clone 3: F09ig0269 {Pbad.rbs.prepro.StrepTag}{<mgfp-5>}{<azo1653 AtD>}(dblterm)
Restriction map of minipreps:
Lanes from left to right: 261-1, 261-2, 263, 265, 269.
Eco/Bam restriction map part sizes:
- F09ig0261: 3039/2069 bp
- F09ig0263: 3039/2069 bp
- F09ig0265: 3039/2096 bp
- F09ig0269: 3039/2660 bp
The gel looks good, so I sent these parts in for sequencing. They are iG049-iG056.
Wednesday, August 12
Checked the strips from yesterday's assembly of internal passengers again and passenger-less final parts. The internal passengers had very small colonies and several were blank. The passenger-less final parts had many colonies. We let the internal passengers incubate for longer and placed the passenger-less set in the fridge to pick colonies later.
Jenn and I started to play detective with parts we have lost along the way: virG, CPG_L6, TshA, and IILK. John's restriction map from 8/1 showed TshA looking fine, but CPG_L6 was missing. We should look at the eco/bam transferred colonies of TshA and the plasmid that was transformed into EC100D 116 pir + (7/31) for CPG_L6.
I miniprepped the some cellulase parts for Patrick:
- pBca9495AC F09ig0144 {Pbad.rbs.prepro.StrepTag}{<cel3A!}{<Pcryo_1225 AtD>}(dblterm) (located in A2 of cellulase stock plate)
- pBca9495AC F09ig0143 {Pbad.rbs.prepro.StrepTag}{<cel3A!}{<Hag AtD>}(dblterm) (located in A6 of cellulase stock plate)
- pBca9495AC F09ig0142 {Pbad.rbs.prepro.StrepTag}{<cel3A!}{<VtaA11 AtD>}(dblterm) (located in A4 of the cellulase stock plate)
- Placed minipreps in iGEM box in Jenn's minifridge.
I transformed these cellulase parts into MC1061 cells and plated on CA antibiotic plates.
I picked colonies (2 clones) of the internal passengers again. Noted which did not have colonies or just one colony to pick:
- A1(0), A5(1), A6(1), A10(0), A12(1), B1(1), B4(1), B5(tiny colonies), B6(tiny), B7(tiny), C1(0), C4(1), C5(0), C6(0), C7(1), C9(1, tiny), C10(0), C11(2nd tiny), C12(0), D2,3,4(0), D5(2nd tiny), D6(1), D7(1), D8(0), D9(1), D10(0), D11(1), D12(2nd tiny), E1, 2(0), F4(0)
I helped get the mgfp assay started with Jenn. Then I picked colonies while she continued to run the assay. The results from the assay were not very promising. The PBS washed left salt crystals on the slides, making it very difficult to detect bacteria. After staining with coomassie blue there were not obvious cell pellets that were stained. There level of surface expression of mgfp may be too low. This is supported by the strep assay results that were pretty much negative across the board for mgfp.
Thursday, August 13
- Spot check yesterday's parts that were picked into 2YT.
Monday, August 31
Induce cultures for tomorrow's AIDA no pass. and mgfp tyrosinase experiments.
Miniprep AIDA no pass in DH10B and send out for sequencing.
Talk to Terry and Jenn about tyrosinase assay. I plan to follow the following protocol tomorrow:
1. Grow up cells with negative controls (1363 induced and tyrosinase treated/ 1363 induced w/out tyrosinase) and positive control (mgfp-AIDA induced and treated with seawater only)
2. Induce cells overnight
3. Pellet cells at 5700 rpm for 10 min. Flick supernatant out in sink.
4. Make 10x solution of tyrosinase enzyme (500ug/ml) in phosphate buffer
5. Add 10 ul of enzyme solution to v-bottom plate except for negative control and positive control wells.
6. Resuspend cells in 100ul of phosphate buffer (two plates with ascorbic acid and two plates without ascorbic acid)
7. Add 90 ul of cell solution to the tyrosinase in the v-bottom plates.
8. Incubate at 25C for 90-180 minutes.
9. Take OD600 reading on the Tecan to observe flocculation.
10. Spin down cells in v-bottom plates at 5500 rpm for 5 minutes.
11. Resuspend cells in seawater or 5% acetic acid (each set of conditions applied to both the ascorbic acid treated plate and the no-ascorbic acid treated plate).
12. Transfer 20 ul to a flat bottom plate (or whatever surface one wants to adhere to).
13. For the 5% acetic acid treated plate, add sodium bicarbonate to bring the solution to a neutral pH.
14. Incubate on surface for 15-30 minutes.
15. Wash with PBS/seawater using 3.14 program on plate washer.
16. Stain with coomassie for 10 minutes.
17. Wash with PBS/seawater.
September
Saturday, September 5
Induce mgfp parts without linkers to incubate with tyrosinase tomorrow and resuspend in seawater as well as acetic acid. Tomorrow's protocol should be as follows:
- Pellet cells in 96 well blocks.
- Resuspend in 100 ul of 0.1 M phosphate buffer.
- Suspend .1 mg of tyrosinase in 2 mL of phosphate buffer.
- In v-bottom 96 well plate, add 10 ul of tyrosinase solution (500mM) and then add 90 ul of cell suspensions to bring tyrosinase concentration to 50mM.
- Incubate at room temperature for 90 minutes.
- Pellet cells in v-bottom plate at 5400 for 8 minutes.
- Resuspend in 100 ul of either seawater or 5% acetic acid.
- Cover with plastic wrapping and let settle overnight.
Also set up uninduced plate of mgfp parts with linkers to Monday. Will induce tomorrow.
Thursday, September 10
- Run BCA assay on 4 mgfp+linkers plates and one mgfp-no linkers plate (all plates were incubated at room temperature overnight in seawater.
- I am hoping to replicate the results from 9/8 with the mgfp+linkers plates. The data from 9/8 showed a lot of positive hits, but there was definitely some background from poor wash of the first plate. Today, I adjusted this wash by running the 3.14 program on a test plate first before proceeding with washes of the actual plates.
Today's assay went as follows:
- Run 3.14 program on washing machine (in corner of room), washing with phosphate buffer (pH 7).
- Add 25 ul of Tris+SDS buffer to each well.
- Set up BCA standards: .25 mg albumin/ml, .75 mg/ml, 1.0 mg/ml.
- Incubate at 50 C for 15 minutes.
- Set up BCA plates for Tecan: add 190 ul of BCA reagents(1:50 ratio of little blue bottle to big white bottle of BCA reagents).
- Take the plates out of the incubator and add 10 ul of the cell lysate to the BCA tecan plates.
- Incubate for 30-40 minutes at 37C.
- Take OD 562 measurements on the Tecan.
- There were no significantly strong signals. The aspiration on the wash robot might be too harsh and remove all of the cells from the surface of the plates. Maybe I should try the flicking technique to remove the supernatant?
BCA Raw data:
File:BCA 9-10 mgfp with and without linkers.xls
BCA data organized: File:BCA 9-10 data organized.xls
Monday, September 14th
Re-ran BCA assay:
- one plate no tyrosinase
- three plates treated with tyrosinase (no ascorbic acid)
- both sets have settled in artificial seawater overnight
---> want to try gentler wash method of "flicking"
- Let incubate for 40 minutes with BCA as usual, did not see significant signals from the tyrosinase-treated plates, so let incubate for 80 minutes to see difference:
Raw data with analysis:
File:BCA 9-14.xls
Raw data with analysis for longer incubation:
File:BCA 9-14 long incubation.xls
- Overall, saw that the plate treated without tyrosinase had significantly stronger signals than those treated with tyrosinase.... very interesting....hmmmm
- Will try to pursue this and run the assay with more replicates.
September 18th
Ran BCA assay for:
- mgfp+linkers
- mgfp-no linkers
- no passengers
- All incubated and settled in artificial seawater
- Each condition ran in 5x
Raw data:
File:9-18 BCA link, no link, no pass.xls
Results analyzed (including t-tests):
File:BCA assay 9-18 results.xls
- Results suggest that linkers enhance the display of mgfp
- Number of hits: mgfp+linkers > mgfp-no linkers > no passengers
September 30th
BCA assay to confirm reproducibility of the:
- mgfp+linkers
- mgfp-no linkers
- displayers only
- Each condition ran in 5x
- Each plate was incubated in artificial seawater.
- Used v-bottom plates like last time
- Use the 'flick' technique again
Raw data:
Media:9-30 BCA data.xls
Data anylized:
Media: BCA 9-30 Results.xls
October
Monday, October 12th
Ran BCA assay in triplicate for to get better quality pictures of the assay for the powerpoint presentation:
- Since we decided to use displayers only as our negative control/point of comparison for the t-tests, 1363 was not included in this set.
Raw data: