Imperial College/Courses/Fall2009/Synthetic Biology (MRes class)/'R' Tutorial/Practical
<html> <body> <script type="text/javascript"> var sc_project=3315864; var sc_invisible=0; var sc_partition=36; var sc_security="8bb2efcd"; </script>
<script type="text/javascript" src="http://www.statcounter.com/counter/counter_xhtml.js"></script><noscript>
</noscript>
</body> </html>
Introduction to 'R'
Part 1: Random Walks
Below is a basic 'R' script to simulate a random walk along the X axis. Random walks are important to model as they relate for example to the phenomenon of diffusion.
- Copy and save the script into a file.
- Run this script within 'R'
<syntax type='R'>
- Random Walk 1D / 1 Trace
-
- number of steps
nbSteps <- 1000
- sampling and path construction
- First method: Looping
- draw from uniform distribtion between [0,1]
x <- runif(n = nbSteps)
- initialise path variable
xSteps <- rep(0, nbSteps) #as opposed to x1Steps <-0
- build independent steps series
for(i in 1:nbSteps){ if(x[i]>0.5){ xSteps[i] <- 1} else{ xSteps[i] <- -1 } }
- build path from independent steps
xPath <- cumsum(xSteps)
- Second method: vectorisation (faster)
- x <- runif(n = nbSteps)
- xSteps <- ifelse(x>0.5, 1, -1)
- xPath <- cumsum(xSteps)
- plot path
plot(xPath, 1:nbSteps, type='l', col='red', lwd =2,
main = 'Random Walk 1D',
xlab = "X coordinate",
ylab = "Steps")
</syntax>
Questions
- Modify this script so that you can overlay as many independent paths as you want (Tip = use the function capability from 'R').
- Write a function to only return the last X coordinate after nbSteps.
- Use the previous function to build the distribution of X coordinates from 1000 independent simulations, after nbSteps.
- Extend the first script to be able to simulate 2D random walks.
Part 2: Enzymatic reaction analysis
In this section, we will be looking at enzymatic reaction data, using a Michaelis-Menten model. The purpose of the 'R' script is to automatise enzyme characterisation from experimental data.
Part 2a: Linear regression analysis
- Copy this script and run it.
<syntax type='R'>
- Enzymatic analysis: Linear regression analysis
concentration <- c(0.3330, 0.1670, 0.0833, 0.0416, 0.0208, 0.0104, 0.0052) velocity <- c(3.636, 3.636, 3.236, 2.666, 2.114, 1.466, 0.866)
df <- data.frame(concentration, velocity) plot(df$concentration, df$velocity)
df$ytrans <- 1/df$velocity df$xtrans <- 1/df$concentration
par(ask=T) plot(df$xtrans, df$ytrans)
lmfit <- lm(ytrans~xtrans, data=df)
summary(lmfit)
coefficients <- coef(lmfit) print(coefficients)
par(ask=T) plot(df$xtrans, df$ytrans) abline(coefficients)
Vmax <- 1/coef(lmfit)[1] Km <- Vmax*coef(lmfit)[2]
print(Vmax) print(Km)
</syntax>
Part 2a:Questions
- Using the help functions in 'R', add comments to the script to detail each command. Use also 'plot' arguments to make plot labels more explicit.
- Modify the script so that experimental data can be read from a file, and analytical results can be exported into a file.
Part 2b: Non-linear regression
In this section, we will use a non-linear regression method to estimate the Michaelis-Menten parameters from the data. The non-linear regression method explored here is the least-square method.
<syntax type'R'>
- Enzymatic analysis: Non-linear regression (Least-square optimisation)
concentration <- c(0.3330, 0.1670, 0.0833, 0.0416, 0.0208, 0.0104, 0.0052) velocity <- c(3.636, 3.636, 3.236, 2.666, 2.114, 1.466, 0.866)
df <- data.frame(concentration, velocity) plot(df$concentration, df$velocity)
nlsfit <- nls(velocity~Vmax*concentration/(Km+concentration),data=df, start=list(Km=0.0166, Vmax=3.852))
summary(nlsfit)
par(ask=T) plot(df$concentration, df$velocity, xlim=c(0,0.4), ylim=c(0,4))
x <- seq(0, 0.4, length=100) y2 <- (coef(nlsfit)["Vmax"]*x)/(coef(nlsfit)["Km"]+x) lines(x, y2)
</syntax>
Part 2:Questions
- As before, using the 'R' help comment this code.
- Modify the code so that the initials values used by the non-linear regression can be read from the parameter file you created in Part 2a.
- Plot on the same graph both fits (linear regression and non-linear regression)
Part 3: Constitutive Gene Expression Modelling
In this section, we will use 'R' to simulate a simple constitutive gene expression model.
The model is given by a system of 2 differential equations to account for the expression of mRNA molecules and Protein molecules:
Following the law of mass action, we can write:
|
The script below uses this model to simulate mRNA and Protein time series. It uses the 'odesolve' package in 'R', and its 'lsoda' function. Check ?lsoda for more details.
<syntax type='R'>
- Gene Expression ODE model
params <- c(k1=0.1, d1=0.05, k2=0.1, d2=0.0025)
times <- seq(from=1, to=3600, by=10)
geneExpressionModel <-function(t, y, p) { dm <- p["k1"] - p["d1"]*y["m"] dp <- y["m"]*p["k2"] - p["d2"]*y["p"] res<-c(dm, dp)
list(res)
}
require(odesolve)
results <- lsoda(c(m=0,p=0),times,geneExpressionModel, params)
print(results)
plot(results[,1], results[,2], xlim=c(0,3800), ylim=c(0,100)) lines(results[,1], results[,3])
</syntax>
Part 3: Questions
- Modify this script so you can read the parameters (k1, k2, d1, d2) from a file, and store the results of the simulation into a file.
- Create a function to return the mRNA and protein steady-state from the parameters (k1, k2, d1, d2).
- Plot the steady-state level of mRNA and protein on top of the simulated results.
- As you can observe, the mRNA level reaches steady-state very quickly in comparison to the protein level. It is therefore possible to use a quasi-steady-state hypothesis on the mRNA, and assume that the level of mRNA is already at steady-state from the start of the simulation. It helps to simplify the model to:
or,
Modify your script to take into account this simplification, and compare both outputs to evaluate how good is the quasi-steady state assumption.
Part 4: Repressed Gene Expression
Very few genes are known to have a purely constitutive expression, most genes have their expression controlled by some outside signals (DNA-binding proteins, Temperature, metabolites, RNA molecules ...). In this section, we will particularly focus on the study of DNA-binding proteins, called transcription factors. These proteins, when binding to a promoter region, can either have an activation effect on the gene (positive control), or a repression effect (negative control). In prokaryotes, control of transcriptional initiation is considered to be the major point of regulation. In this part of the tutorial, we investigate one of the most common model used to describe this type of interactions.
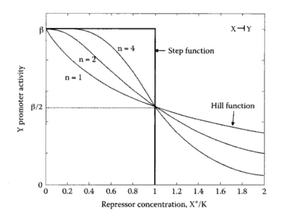
Let's first consider the case of a transcription factor acting as a repressor. A repressor will bind to the DNA so that it prevents the initiation of transcription. Typically, we expect the transcription rate to decrease as the concentration of repressor increases. A very useful family of functions to describe this effect is the Hill function:
[math]\displaystyle{ f(R)=\frac{\beta.{K_m}^n}{{K_m}^n+R^n} }[/math].
The Hill function can be derived from considering the transcription factor binding/unbinding to the promoter region to be at equilibrium (similar to the enzyme-substrate assumption in the Michaelis-Menten formula).
This function has 3 parameters: [math]\displaystyle{ \beta, n, K_m }[/math]:
- [math]\displaystyle{ \beta }[/math] is the maximal expression rate when there is no repressor, i.e. [math]\displaystyle{ f(R=0)=\beta }[/math].
- [math]\displaystyle{ K_m }[/math] is the repression coefficient (units of concentration), it is equal to the concentration of repressor needed to repressed by 50% the overall expression, i.e [math]\displaystyle{ f(K_m)=\frac{\beta}{2} }[/math]
- [math]\displaystyle{ n }[/math] is the Hill Coefficient. It controls the steepness of the switch between no-repression to full-repression.
|
Hill function for transcriptional repression:
|
Following the law of mass action, we can write the following ODE model:
Part 4:Questions
- Write a 'R' script to simulate the mRNA and Protein expression as a function of the repressor concentration.
- We want to plot the transfer function between the repressor [R] concentration and the protein steady-state level.
- Suggest an application where this genetic circuit might be useful.