ISEEM Progress March 2009
| Home | Project | People | News | For Team | Calendar | Library |
I. Progress made toward completion of our goals
A. Personnel
- There have been no personnel changes in the Pollard lab.
- Jessica Bryant began working as a technician on the iSEEM project in the Green lab.
- Martin Wu has left the Eisen lab for a faculty position at U. Virginia. We are in the process of making a subcontract to him.
- Sourav Chatterji left the Eisen lab for a industry position.
- The Eisen lab is interviewing potential replacements for Sourav Chatterji and have identified a candidate, Morgan Languille from Fiona Brinkman's lab. We are working on getting him an offer.
B. Research
Eisen lab
We’ve identified 66 gene families that spread across at lease 90% of the 85 bacterial genomes with even distributions. As gene marker candidates, we’ve identified 27 families that are single copied in all the organisms. 25 families have multiple copies of genes because of recent gene duplications, which were suggested by phylogenetic trees. 27 of the 52 families were already included in AMPHORA as phylogenetic markers. Maximum likelihod trees were built for the non-redundant sequences of the 52 gene families, as well as for the small subunit rRNAs and the concatenated alignments of 31 AMPHORA protein markers. Topological distances between the individual gene trees and the concatenated genome tree were calculated by the TOPD/FMTS package. Both NODAL and SPLIT distance results suggested that the 25 new phylogenetic maker candidates are as good as the 27 AMPHORA makers if we only take the similarity of gene trees and the genome tree into account. More detailed comparison is underway to estimate how the new markers perform for different taxonomic groups at different taxonomic levels.
We are also developing a new pipeline, in similar lines of AMPHORA, for direct identification and phylogenetic analysis of metagenomic nucleotide data. The new pipeline works by identifying marker sequences in the metagenomic data using HMMsearch and create a multiple alignment of identified marker regions using Muscle. It thus produces untrimmed, unmasked alignments, which are then reverse translated by overlaying nucleotide data on the peptide sequence alignments. Thus generated nucleotide marker alignments which can then be studied for various attributes, such as evolutionary patterns and evolutionary rates. Some of the salient feature of this new pipeline include parallel processing for cluster computing, incorporation of nucleotide read in sync with the peptide data and ability generate untrimmed and unmasked alignment, thus overcoming the problem of omission of sequence data. As of now, the pipeline has been built upto alignment generation and is now being further extended to include supertree/supermatrix based phylogenetic tree creation taking account of missing data in the metagenomic reads. The pipeline will also be extended for additional markers.
Green lab
Metagenomic analysis of community phylogenetic structure
We developed a pipeline to analyze the phylogenetic structure of microbial communities based on metagenomic data. AMPHORA is used to identify sequences from 31 gene families in a metagenomic data set. For each gene family, the aligned metagenomic sequences are combined with the AMPHORA reference alignment of ~500 sequences for each gene family. The AMPHORA reference sequences serve as a phylogenetic anchor or scaffold to improve resolution of phylogenetic relationships among metagenomic sequences. A phylogenetic tree is inferred from the combined metagenomic and reference sequences using maximum likelihood methods. The reference sequences are then pruned from the tree, leaving a phylogeny linking the metagenomic sequences. This phylogenetic tree is then used to estimate several measures of community phylogenetic structure within and among environmental samples using the picante R package.
We applied this pipeline to the HOT/ALOHA metagenomic data set as a proof of concept. Phylogenetic diversity varied predictably along enviornmental gradients in this data set, unlike taxonomic diversity measured using 16S rRNA OTUs. However, patterns of phylogenetic diversity differed among the 31 gene families we examined. It is unclear whether this variation in phylogenetic diversity among gene families is a sampling issue or reflects underlying variation in selection on different gene families along environmental gradients. Our plans for this deliverable are to explore other means of inferring phylogenetic relationships among sequences such as using a concatenated alignment, applying this method to a wider range of gene families as they are added by the Eisen lab (see above), and to apply these methods to larger metagenomic data sets available on CAMERA to determine whether the paterns we observed are general.

Novel Biodiversity Measures
We are continuing to develop theoretical predictions for phylogenetic alpha- and beta- diversity. As a first step, we have developed a spatially-explicit, neutral (that is, non-interacting) model for community taxonomic diversity. The approach of focussing on taxonomic diversity in the first instance has three advantages. First, there is a synergy with related approaches in the Pollard lab. Second, the taxonomic model provides a basis on which to later build phylogenetic structure. And thirdly, it will allow us quickly to make contact with data.
We have derived a functional differential equation for the spatial community partition function, Z[H]:
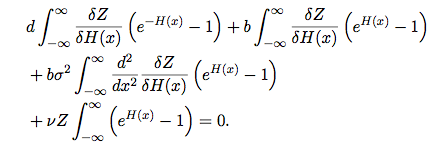
The parameters in this equation, b, d, [math]\displaystyle{ \sigma }[/math] and [math]\displaystyle{ \nu }[/math] represent birth rate, mortality rate, dispersal distance and speciation, respectively. While the significance of Z[H] is not intuitively obvious, finding its solution will allow us to make neutral predictions for a wide range of community distributions, including the Species-Area Relationship and the spatial scaling of the Species Abundance Distribution. The agreement or deviation of microbial data from these predictions will inform us how well or badly the assumption of neutrality performs, and will guide the inclusion of additional complexity to our model.
The approach we have taken to solving this formidable equation is to look for a partial, cut-down solution, for the function [math]\displaystyle{ z_{SAD}(h,A) }[/math]. This function is simpler than the whole partition function, [math]\displaystyle{ Z[H] }[/math], but generates the moments of the Species Abundance Distribution, and how this distribution scales with area, [math]\displaystyle{ A }[/math]. We have reduced our master equation to a set of partial differential equations describing [math]\displaystyle{ z_{SAD} }[/math], and this step allows us to focus on one key measure of taxonomic community diversity, while significantly reducing the difficulty of the equations we need to solve. Our next step is to solve these equations to obtain a neutral prediction for the spatial scaling of the Species Abundance Distribution.
Pollard Lab
Species-Area Relationships
A fundamental question in microbial community ecology is how taxonomic richness scales with area. Answering this question has implications for understanding microbial evolution – whether microbes are primarily adapted to specific niches or cosmopolitan – and estimating regional to global microbial diversity. The studies that have examined this question have yielded widely divergent estimates of z (the exponent in the power function that is often used to model taxa-area relationships) with values ranging from less than 0.05 to greater than 0.4. For non-microbial communities, values of z are generally greater than 0.2, so the low estimates are noteworthy. We believe that the low values of z for microbes result from using biased estimators rather than a difference between microbal and macroorganismal biology.
The scaling parameter in taxa-area relationships can most directly be estimated by plotting taxonomic richness against area and fitting a power function via nonlinear regression (or linear regression with log-transformed observations). However, estimating z for microbes is confounded by the fact that it can be difficult to census microbes at large spatial scales. As a result, studies have implemented a distance-decay approach to estimating z. In this approach, one censuses communities in small plots of equal area or volume. The parameter z is then estimated by examining how the similarity between pairs of communities decreases with the distance separating them. In three recent microbial studies (Green et al 2004, Horner-Devine et al 2004, Fierer & Jackson 2006), an estimator of z derived by Harte & Kinzig (1997) was applied and estimates of z were less than 0.1.
We developed a novel distance-decay based estimator of z that utilizes a logistic regression model (rather than linear) and is based on different statistical theory than the Harte & Kinzig estimator. Our estimator is based on minimal assumptions about the data and is robust to deviations from these assumptions.
We assessed our method using six ecological data sets (of plants, animals and microbes) where z could be estimated directly from richness versus area curves as well as using distance-decay. Our estimator agrees remarkably well with the richness-area based estimator (which is known to be unbiased). The Harte & Kinzig estimator, in contrast, is severely biased.
We are currently extending our results to develop a more general estimator and to assess its performance in simulations. For metagenomics applications, we are particularly interested in how our method performs when not all taxa that are present are observed in a sample (i.e. sequencing depth is not sufficient to detect the rarer taxa).
C. Communications, Collaboration, Outreach and Education
Publications supported by this grant
- T. Woyke, G. Xie, A. Copeland, J.M. González, C. Han, H. Kiss, J.H. Saw, P. Senin, C. Yang, S. Chatterji, J-F. Cheng, J. A. Eisen, M. E. Sieracki and R. Stepanauskas. Assembling the marine metagenome, one cell at a time. In press. PLoS One.
- O'Dwyer, J., Lake, J., Ostling, A., Savage, V., Green, J.L. (2009). An integrative framework for stochastic, size-structured community assembly. 2009. Proceedings of the National Academy of Sciences, Online Early (doi: 10.1073/pnas.0813041106).
New collaborations
- Katie Pollard met with Joe DeRisi and his lab. We are looking into collaborating on a project to identify novel phylotypes and/or genes in the virus domain.
- Multiple iSEEM personnel are collaborating with Erck Matsen and Robin Kodner on phylogenetic informatics
- Jonathan Eisen is beginning to work with multiple faculty at UC Davis on methods for phylogenetic informatics
- Jonathan Eisen has been working with multiple labs at UC Davis in order to help them develop projects and proposals in the area of microbial diversity studies. This includes collaborations with Artyom Kopp (Drosophila gut microbes see http://www.eve.ucdavis.edu/kopplab/), Venkatesan Sundaresan (Rice Root Microbiome see http://www-plb.ucdavis.edu/Labs/sundar/), Kate Scow (affect of elevanted CO2 on soil microbes http://scowlab.lawr.ucdavis.edu/), and multiple others. In these cases, the existence of the iSEEM project has helped convince labs that there is enough local expertise in metagenomics to start to use this technology.
- Jonathan Eisen has subbmitted two proposals to acquire a Roche GS Titanium system for microbial diversity studies at Davis. As with the collaborations above, the existence of the iSEEM project was a driving force behind the development of these proposals.
Talks
- Steven Kembel presented iSEEM research in an invited talk "Ecology without species: phylogenetic perspectives on microbial diversity" at the University of Michigan Early Career Scientists Symposium.
- Jessica Green gave a talk in Greece on iSEEM research at the University of Ioannina
- Jonathan Eisen talked at the JGI User Meeting including some results from this project.
- Jonathan Eisen gave the UC Davis Evolution and Ecology Seminar Series talk on Feb 5, 2009 and discussed the iSEEM project
Teaching
- Katie Pollard is teaching a course at UCSF (Spring quarter) on analysis of genomic data using R.
- Jonathan Eisen discussed phylogenetic metagenomics in a lecture for the UC Davis Bodega Bay Workshop on Phylogenetics
- Jessica Green discussed phylogenetic metagenomics in her undergraduate course Biodiversity at the University of Oregon (Winter quarter)
Outreach
- Jonathan Eisen gave a "public" talk at UC Davis in honor of Darwin Day 2009 and discussed in part the uses of phylogeny in studying uncultured microbes
- Jonathan Eisen and Thomas Sharpton "live blogged" the JGI User Meeting (https://friendfeed.com/rooms/jgi-user-meeting) and discussed in the live blog many aspects of metagenomics
- Jonathan Eisen is featured in an article on Metagenomics in Genome Technology's April Issues (http://www.genomeweb.com/metagenomics-move)
II. Group meetings
- Notes for biweekly Group meetings are attached as PDF files.
- Notes for biweekly PI meetings are also attached as PDF files.
- Other meetings
- Robin Kodner and Erick Matsen visiting the Eisen lab to discuss phylogenetic metagenomics
III. Any unexpected challenges that imperil successful completion of the Outcome
The #1 concern is still in our interactions with CAMERA. We are in on/off discussions with them about the implementation of our software and these discussions are proceeding but we still do not have any actual products up and running at CAMERA as far as we are aware. In addition we still do not have access to computational resources (e.g., cluster time) through CAMERA. We probably need some help from GBMF in order to progress more rapidly in terms of making use of CAMERA resources when appropriate and getting our algorithms and tools available to the community through CAMERA.
