ISEEM AnnualReport 2008
| Home | Project | People | News | For Team | Calendar | Library |
Annual report 2008
I. Progress made toward completion of our goals
A. Personnel
People directly supported by the grant are:
- Jessica Green
- Steven Kembel, PostDoc
- James O' Dwyer, PostDoc
- Liz Perry, PhD Student
- Jess Bryant, Research Assistant
- Katie Pollard
- Josh Ladau, PostDoc
- Sam Riesenfeld, PostDoc
- Jonathan Eisen
- Dongying Wu, Staff Scientist
- Martin Wu, Staff Scientist. Note Martin has moved to a faculty position at U. Virginia. We are working on having a subcontract to him to continue work on the project.
- Srijak Bhatnagar, Bioinformatics Engineer
- Marisano James, PhD student, Population Biology Graduate Group
- Aaron Darling, PostDoc. He will begin work on the project in June 2009.
- Sourav Chatterji, PostDoc for Year 1. Has now moved to another job.
Others involved in some components of the project but not directly supported by the grant include Amber Hartman and Jenna Morgan, (PhD students at UC Davis).
B. Research progress organized in terms of grant deliverables
1.1 Guidelines and weightings for using different gene families in metagenomic based diversity assays.
1.1a. Generate alignments and trees for targeted set of 50-100 gene families; Deliver trees to projects 1b, 1c, 1d, 2a,and 2b; Generate scores for targeted set of 50-100 gene families; Test utility of scores using simulated and real metagenomic data sets.
- Target date for completion 12/2008.
- Status - 90% Completed
Generate alignments and trees for targeted set of 50-100 gene families
Status - effectively complete.
A key step in many metagenomic projects is making multiple sequence alignments of genes or the proteins they encode. Usually in such alignments one wants to include sequences from multiple sources (e.g., PCR, genomes) in addition to metagenomic data. Such multiple sequence alignments are central to many of the subprojects that make up our iSEEM project.
Unfortunately, to make full use of multiple sequence alignments for our and other projects, it was not possible to use existing software tools. There were two key limitations to existing tools. First, many require too much manual examination of the results to be used in a high throughput manner to analyze the ever increasing number and size of metagenomic data sets. Second, even when alignments were considered to be high quality overall, certain regions of the alignments were not high quality.
Therefore we needed to design a method that could both be fully automated and could identify or weed out regions of the alignment that were of poor quality. We have now developed exactly such a method, which we have called AMPHORA (an acronym for AutoMated PHylogenOmic infeRence Algorithm). A paper describing the method was published in 2008 (Wu and Eisen, 2008; see http://genomebiology.com/2008/9/10/R151) and the software has been made freely available. In summary this method works through the creation and use of curated multiple sequence alignments for protein families of interest. From these alignments we make Hidden Markov Models (HMMs) which are a statistical representation of the sequence patterns seen in the multiple alignments. In addition, we label specific regions of the alignments as being high or low quality using something known as a sequence "mask." With these two things (a HMM for a protein family and a mask) we can then generate high automated alignments of members of that protein family and also identify regions of the alignment that are high quality. We showed in our paper that this can then be used for analysis of both genome data and metagenomic data. For this paper we generated curated HMMS for 31 protein families.
Using AMPHORA we have now been generating multiple sequence alignment for these 31 protein families from multiple metagenomic data sets (e.g., in the paper we focused on the Sargasso Sea data from Venter et al. 2004). For the iSEEM project we have decided to focus some initial analyses and methods development on a specific metagenomic data set - the ALOHA depth series data (Delong et al. 2006). We have generated alignments for the 31 protein families from this data set and have made them available to all iSEEM members.
In making these alignments we have come to the realization that we also need to make alignments of the DNA encoding these proteins and not just the amino acids. We are currently working on automating the "back alignment" so that we can align the proteins (i.e., the amino acids, which allow for more accurate alignments) and to then capture the DNA for each protein, keeping the alignment in register. We are focusing on this rather than on expanding the marker gene list from 31 to > 50. However, in a separate activity, we have designed an automated method (called ZORRO, see below) which allows us to make use of any new protein family of interest with no manual curation effort (manual curation was required for the 31 marker genes).
Deliver trees to projects 1b, 1c, 1d, 2a,and 2b
- Target date for completion: 12/2008
- Status - delayed a few months due to need to determine which phylogenetic methods to use. Expected completion 4/2009.
We have built evolutionary trees for all the proteins corresponding to the 31 marker genes from the ALOHA data set. For this we used the maximum parsimony method of RAXML as we used in the paper describing AMPHORA (Wu et al. 2008). However, it is unclear if this is the ideal phylogenetic reconstruction method to use for this type of analysis. The difficulty is that metagenomic data is fragmented and we would like to include many fragments in a single phylogenetic tree. What method to use to do this is unclear. We have begun to investigate this by both surveying the literature and asking the global phylogenetic research community. To survey the community we sent emails to 10+ phylogeneticists and posted questions on Jonathan Eisen's blog (see http://phylogenomics.blogspot.com/2008/01/calling-all-phylogeneticists-we-need.html). We have collected this and other information on the project Wiki (http://iseem.private.openwetware.org/wiki/Phylogenetic_Methods) and continue to gather information. Although there are some excellent ideas and methods, it seems to us that nobody has truly tested the diverse array of possible methods for how well they work with metagenomic data. Therefore we are pursuing this as a new spin-off subproject within the iSEEM project.
Several possible methods include supertrees, supermatrices, and a variety of distance matrix based methods that use different subalignments (without too many gaps) to generate different pairwise distances based on the amount of overlap between the pair of reads. Samantha Riesenfeld and Steve Kembel are working on this evaluation of phylogenomic methods with significant input and support from Srijak Bhatnagar and Martin Wu. In order to evaluate these different tree building approaches, we are developing a simulation pipeline, which will most likely be based on the MetaSim program described in a paper by Richter et al [1]. Steve Kembel and Samantha Riesenfeld are taking a lead on these simulations. The wiki describes our ideas and approaches: http://iseem.private.openwetware.org/wiki/Simulations_of_Metagenomic_Data.
While we have been researching phylogenetic methods to use, we have been working on building a relational database to capture and store and share the information about phylogenetic trees, alignments and other aspects of protein families being studies. For this and other work, we are making use of a genome database developed by Martin Wu in the Eisen lab called ComboDB. ComboDB is a comprehensive database of complete genome sequences (including information on proteins, the genes encoding these proteins (CDSs), rRNAs, tRNAs, and genome assemblies) and their various attributes, such as taxonomy, GC content, gene locus, etc). In the past this database has been stored in a flat file format. This database has now been converted into a hybrid database, where the various information will be stored as a relational database and the sequences as a flat-files linked to this relational database. Such a database with add enhanced querying, easy accessibility and better interface for retrieval and mining of data. In essence, it will house the data used in various genomic analysis to develop the parameters for metagenomic analysis and will link to many tools for on-portal analysis, in an easy to access fashion. As of now, the database has been created and the front-end for it is under construction, following which various tools will be added to it.
Generate scores for targeted set of 50-100 gene families
- Target date for completion: 12/2008
- Status: Completed.
In order to identify potential phylogenetic markers for metagenomic studies, we are developing methods to analyze the gene distributions across genomes with compete genome sequences. We selected 85 bacterial and 15 archaeal genomes for the initial study. A maximum likelihood tree of 720 bacterial genomes based upon 31 concatenated genome markers were used for the bacterial genome selection, while the selection of archeal genome was based on a tree of the archeal radA genes. A program called maxPD was developed to automatically select taxa from a phylogenetic tree so the taxa are listed according to their contributions to the phylogenetic diversity based on the tree. The genomes that contributions most phylogenetic diversity were selected. In the selecting process, we only selected the genomes with publications and we also avoided genomes that undergone severe genome reductions.
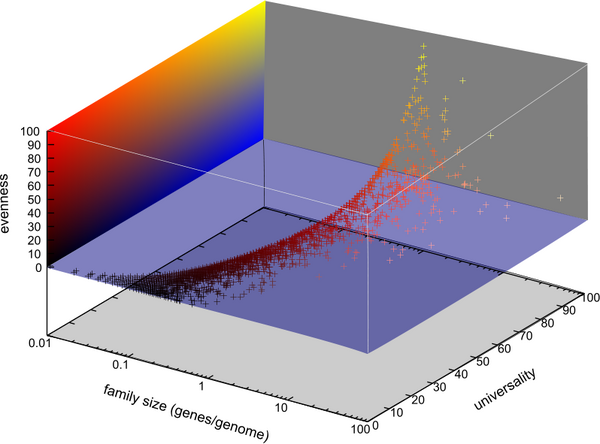
A protocol has been developed to classify gene families automatically. The protocol is base upon Blastall and the Markov clustering algorithm so the protocol are automated, robust. We use an e value cutoff of 1e-10 in our test run. For the 313139 genes from the 100 selected genomes, we identify 23336 gene families that include a total of 239453 genes. In order for automatically select gene families for potential markers, we've developed a program that not only evaluates the universality of the gene families, but also estimates the evenness of the distributions of the genes across a selected group of genomes (See the figure below).
Across all 100 the selected genomes, we identified 30 gene families that appear in almost all the genomes with even gene distributions, 12 were ribosomal protein subunits that were already included in our gene marker collections which indicate the approach works in principal. The rest of the candidates are mostly tRNA synthetases among others. Different cutoffs will be applied for more potential marker selection, and the evenness and distribution estimation program was developed in a way that potential markers for specific phylogenetic groups (for example, a phylum or a class) can be easily identified.
Test utility of scores using simulated and real metagenomic data sets.
- Target date for completion: 12/2008
- Status: In progress. Expected completion 4/2009.
This work is in progress and was delayed slightly due to the need for design of better metagenomic simulation tools (see above).
1.1b. Generate alignment, trees, and scores for expanded set of ~500 families; Test utility of these scores using simulated and real metagenomic data sets.
- Target date for completion 12/2009.
- Status: Ahead of schedule.
Generate alignment, trees, and scores for expanded set of ~500 families
- Target date for completion 12/2009.
- Status: Ahead of schedule. Expected completion 6/2009
One of the challenges for the iSEEM project and an project attempting to carry out phylogenetic analysis of many different genes is the need to assess the quality of multiple sequence alignments that are used for the phylogenetic reconstructions. In the past, including in the development of the STAP and AMPHORA software outlined above, we have carried out extensive manual examination of alignments to identify regions of the alignment that may not be useful for phylogenetic analysis. Known as "masking" this manual work takes both significant expertise and time.
We have developed ZORRO, a probabilistic masking program for taking into account alignment reliability during phylogenetic inference. Using posterior probabilities of a pair HMM based alignment model, ZORRO assigns confidence scores to each column in the alignment. Using either a cutoff or weighting based approach, these confidence scores can be seamlessly integrated into any phylogenetic inference pipeline.
Using the standard Balibase benchmark data-set, we were able to demonstrate that ZORRO is able to measure alignment accuracy with very high (>95%) sensitivity as well as specificity. We have also created a "benchmark" data-set of Alphaproteobacteria genes for comparing phylogenetic inference protocols in bacteria. Using this benchmark data-set, we were able to demonstrate that phylogenetic protocols using ZORRO showed statistically significant improvement over protocols not using any masking.
Test utility of these scores using simulated and real metagenomic data sets
- Target date for completion 12/2009.
- Status: Expected completion 12/2009
Work on this will begin once the scores are determined as described above.
1.1c. Integrate scores with development of diversity assays.
- Target date for completion 11/2010.
- Status: Expected completion 11/2010.
1.1d. Create and update database of scores for different genes and integrate database into CAMERA.
- Target date for completion 11/2010.
- Status: Expected completion 11/2010.
1.2 Searching for novel phylogenetic types in metagenomic data
1.2a. Develop an automated system for novel branches using rRNA sequences in metagenomic data.
- Target date for completion 12/2008.
- Status: Complete.
A paper describing and automated system for phylogenetic analysis of rRNA sequences (which could come from any source including PCR or metagneomic projects) was published July 2, 2008: Wu D, Hartman A, Ward N, Eisen JA (2008) An Automated Phylogenetic Tree-Based Small Subunit rRNA Taxonomy and Alignment Pipeline (STAP). PLoS ONE 3(7): e2566.
The paper describes software for performing automated alignments and phylogenetic analyses of rRNA sequences. The software was developed over many years in the Eisen lab and the final work on it was supported by this project. It will be used as a part of the rRNA analyses to be carried out for this project. In the paper we also describe a method for searching for novel phylogenetic types using rRNA data.
1.2b Integrate methods into CAMERA.
- Target date for completion 12/2009.
- Status: Expected completion 12/2009.
We are working with CAMERA to convert our STAP rRNA analysis pipeline (which was described in the previous report) into a Kepler Workflow to be runnable from within CAMERA. We had a meeting with the CAMERA and Kepler groups to plan how to tackle this and work is underway to convert our initial Kepler Workflow into something that can be used by CAMERA.
1.2c. Develop an automated system for searching for novel branches for protein coding genes.
- Target date for completion 12/2009.
- Status: Expected completion 12/2009.
Preliminary work has begun on this approach as an extension of the methods described in the rRNA "STAP" paper discussed above. A manuscript on the first application of this approach is being readied for submission where we searched for novel phylogenetic types using two protein coding genes: recA and rpoB. The work will be extended to apply to all protein families.
1.2d. Further novelty activities. Identify novel branches for each gene family; Identify other genes linked to genes identified as novel; Develop a Unifrac-like system for identifying the amount of novelty in metagenomic data for a particular gene family; Compare the novelty of phylogenetic marker gene families versus functionally important families; Compare analysis of gene family novelty with metagenomic sample metadata to determine if any types of environments are enriched for novelty.
- Target date for completion 11/2010.
- Status: Expected completion 11/2010.
1.2e. Integrate methods into CAMERA.
- Target date for completion 11/2010.
- Status: Expected completion 11/2010.
1.3 Metagenomic analysis of community phylogenetic structure
Traditional approaches to understanding biological diversity have largely focused on taxonomic diversity, the number and relative abundance of organisms from different taxonomic units such as species. These measures of diversity ignore an important aspect of biological diversity, the evolutionary or phylogenetic relationships that link organisms. It may be challenging to estimate the richness of taxonomic units such as species in metagenomic data sets, given the fragmentary nature of the data, the difficulty of assigning individual fragments to taxonomic groups, and the more general issue of how to define species or OTUs in microbial communities given variation in evolutionary rates among organisms and gene families. An understanding of community phylogenetic diversity offers the potential to avoid some of these difficulties, and to understand not just how many kinds of organisms coexist in an environment, but who those organisms are, and where they have come from on the tree of life. Ultimately, these approaches may allow us to understand the processes that drive and maintain biological diversity in different environments, but phylogenetic diversity measures have not yet been developed or applied to metagenomic data.
We will identify existing methods or develop new methods that are able to quantify phylogenetic diversity given challenges such as the fragmentary nature of metagenomic data, the potential difficulties with phylogenetic inference in these data sets (section 1.1a), and the difficulty of identifying the source organism of individual sequence fragments. Our general objectives are to quantify various aspects of phylogenetic diversity in metagenomic data sets, including estimate the relative phylogenetic diversity among and within communities, estimate relative evolutionary differentiation along environmental gradients, and to reconstruct the evolutionary history of habitat associations in microbes. Examples of specific methods that will be applied include measures of the total evolutionary history represented in different samples, pairwise phylogenetic distances within and among communities, measures analagous to Fst and Gst statistics developed in population genetics, and measures similar to traditional taxonomic diversity measures such as Simpson or Shannon diversity but incorporating information about evolutionary relationships among taxa. We will evaluate the performance of these different methods and compare observed patterns of phylogenetic diversity to the predictions from simulations and theoretical models of community assembly and evolutionary diversification (see section 1.4b).
1.3a. Metagenomic community phylogenetic analysis of rRNA genes.
- Target date for completion 12/2008.
- Status: 75% complete.
Our initial approach to understanding the phylogenetic diversity in metagenomic data sets has focused on using rRNA genes to measure phylogenetic relatedness among co-occurring organisms in these data sets. SSU rRNA genes were identified both from PCR sequencing of 16S/18S genes from environmental samples (i.e. 16S sequences sequenced from DeLong and GOS samples and available on CAMERA), as well as through scans for genes from these families in metagenomic shotgun sequencing datasets using STAP software developed by the Eisen lab. Phylogenetic trees linking individual sequences in the metagenomic data sets were then constructed using neighbor joining and likelihood methods. Novel phylogenetic inference methods for metagenomic data will be applied to rRNA alignments as they are tested and developed (sections 1.1 and 2.1).
Analyses of phylogenetic diversity based on rRNA genes in the DeLong data set indicate that taxonomic diversity (number of OTUs) and phylogenetic diversity (phylogenetic distances separating sequences) varied independently. Taxonomic diversity based on a range of similarity cutoffs to define OTUs varied relatively little among samples arranged along a water depth and environmental gradient in this data set. Conversely, phylogenetic diversity was strongly correlated with environmental conditions in samples, and samples that were more similar in terms of environment also contained sequences separated by small molecular distances. Randomization tests identified clades that were positively and negatively associated with particular environmental conditions at different depths. These habitat associations arose relatively deep in the tree of life, which explained how phylogenetic diversity but not taxonomic diversity varied predictably among habitats: most samples contained similar numbers of OTUs, but samples contained phylogenetically distinct assemblages. With increasing depth, samples tended to contain sequences from distinct portions of the tree of life, and individual sequences increasingly tended to not occur with other closely related sequences, leading to an increase in phylogenetic diversity with depth while taxonomic diversity remained relatively constant.
Remaining work for this deliverable includes applying these methods to other public metagenomic data sets, comparison of the performance of different phylogenetic and taxonomic diversity metrics, methods of gene identification, and tree inference methods on conclusions about microbial diversity in metagenomic samples, comparison of observed patterns with predicted patterns under various ecological and evolutionary models, and preparation of a manuscript describing the results of this work.
1.3b. Metagenomic community phylogenetic analysis of all 50 gene families identified in 1a
- Target date for completion 11/2010.
- Status: Expected completion 11/2010.
We will estimate phylogenetic diversity in metagenomic data sets based on the phylogenies for 50 functional gene families developed in step 1a (phylogenetic trees for 50 gene families) using the methods described in step 1.3a.
1.3c. Assess utility of gene families from community phylogenetic perspective.
- Target date for completion 11/2010.
- Status: Expected completion 11/2010.
We will combine all methods and results from steps 1.3a and 1.3b to assess the relative utility and performance of different gene families to measure community phylogenetic structure in metagenomic data. We will determine the relative variability of different gene families among and within metagenomic samples, and link differences in phylogenetic diversity of gene families to functional annotations of those genes, in order to understand the relative importance of different processes and environmental gradients on the structure of microbial assemblages in different habitats.
1.3d. Integrate methods into CAMERA tools.
- Target date for completion 11/2010.
- Status: Expected completion 11/2010.
All community metagenomic phylogenetic diversity methods will be implemented and made available in open-source software packages (i.e. Phylocom: Webb et al. 2008, Picante: Kembel et al 2008) and we will work with CAMERA to integrate these methods into their online tools.
1.4. Estimating biodiversity from metagenomic samples
Quantifying the richness - or number - of operational taxonomic units (OTUs) in biological communities has been the gold standard of microbial biodiversity research (e.g. Quince et al. 2008, Shaw et al. 2008). However for macro- and microorganisms alike, this focus is changing. There is growing interest in quantifying not only the number of taxa in communities, but also the phylogenetic relatedness of those taxa and the traits those taxa possess [see attached Green lab publications Bryant et al. 2008, Green et al. 2008]. Here we describe our assessment of traditional OTU richness estimators as applied to metagenomic data (section 1.4a). We then outline our progress towards the development of novel approaches for both characterizing and understanding microbial metagenomic diversity (section 1.4b).
1.4a. Assess the fidelity of currently used species (or phylotpye) richness estimators
- Target date for completion 12/2008.
- Status: 80% complete.
To assess the fidelity of currently used OTU richness estimators, we analyzed the 16S sequence data available on CAMERA from the DeLong Hawaii Ocean Depth-series (discussed in Section 1.3). At each depth, we calculated four commonly used OTU richness estimators: the Chao1, Jacknife, ACE, and boostrap. We used DOTUR [2] to calculate all diversity statistics. Our results corroborate those reported in Shaw et al. 2008, namely that all statistics yielded qualitatively similar diversity rankings of the samples for a given sequence similarity cut-off. Additionally, all the estimators gave qualitatively similar estimates of total richness. The one exception to this pattern was the bootstrap statistic, which appeared to be an outlier in that it consistently estimated substantially lower OTU richness relative to the other diversity estimators, a trend that amplified at higher sequence similarity cut-off values.
In isolation, these preliminary results suggest that the majority of OTU richness estimators are useful for ranking microbial diversity among metagenomic samples. But the fidelity of these estimators becomes questionable when considered in tandem with phylogenetic diversity measured across the same samples. A comparison of Figure (1.4a, left) and Figure (1.3a TD/PD-depth, above) shows that both the observed and estimated OTU richness varied unpredictably along the depth gradient, whereas phylogenetic diversity systematically increased with depth and was strongly correlated with environmental conditions. These findings, and those discussed further in Section 1.3a, substantiate our current focus on contemporary measures of phylogenetic diversity, and also the development of novel metagenomic diversity measures that incorporate information on phylogenetic relatedness (see Section 1.4b). However, given that taxonomic diversity estimators are a widely used benchmark for comparative analysis of microbial diversity, we aim to incorporate our findings into a forthcoming manuscript focused on phylogenetic diversity. Thus, as a component of the research outcome described for Section 1.3a, remaining work for this deliverable includes applying these methods to other public metagenomic data sets, comparison of the performance of different phylogenetic and taxonomic diversity metrics, and preparation of a manuscript describing the results of this work.
1.4b. Develop and evaluate novel biodiversity estimators
- Target date for completion 11/2010.
- Status: 40% complete. Expected completion before 11/2010.
What factors drive community assembly? This is one of the most important and debated topics in theoretical ecology, and to answer this question we are developing theoretical predictions for novel measures of biodiversity. The progress made so far in the Green Lab has led to a slight change in focus on topic 1.4b; the advent of environmental metagenomics offers an unprecedented opportunity to develop and test microbial ecological theory, which will in turn inform the development of measures and estimators of biodiversity. Our theoretical models will go beyond taxonomic diversity to make novel predictions for phylogenetic alpha- and beta-diversity, and the spatially-explicit data from the Global Ocean Survey dataset will be a crucial tool in developing these models, providing a stringent evaluation of their results.
We have focused on two related strands of theory, both intended to meet the unique challenges posed by metagenomic data. The first is a phylogenetic development of the ecological beta-diversity index F(r), which was first introduced in ecology in the context of Neutral Biodiversity Theory (NBT) (Chave and Leigh). NBT is a parsimonious theory which emphasizes stochastic, random forces in community assembly, and F(r) is the neutral prediction for the probability that two individuals in a community, separated by distance r, are of the same species. In collaboration with Helene Morlon, we will draw on tools from population genetics and coalescent theory to generate predictions for a new metric, F(r,s), the probability that two individuals separated by distance r have phylogenetic relatedness, s. We will test our model using the phylogenetic analyses being developed by other members of the iSEEM group, and the agreement or departure of metagenomic data from neutral predictions will be a crucial guide in understanding the processes underlying microbial community structure.
Our second project also has a neutral flavor, but will extend the predictions for F(r,s) to more general measures of phylogenetic beta-diversity (Bryant et al, 2008). The Green Lab has already derived novel mathematical models, which draw on the functional methods of quantum field theory to make neutral ecological predictions. The first example of this kind of `ecological quantum field theory' integrates the effects of individual body-size and neutral stochasticity, both of which are known to strongly influence community assembly (West Brown and Enquist, Hubbell), and is currently in press (O'Dwyer et al 2009 File:Stochastic size manuscript revised3.pdf). Our main ongoing focus will be on developing a spatially-structured version of this model, and we will use its solutions in conjunction with the framework of Morlon et al. 2008 to make general predictions for ecological beta-diversity.
1.4c. Apply diversity estimators to public data
- Target date for completion 11/2010.
- Status: Expected completion 11/2010.
2.1 Molecular evolution of gene families
2.1a. Develop and evaluate molecular evolutionary methods for metagenomics
- Target date for completion 12/2008.
- Status: 75% complete..
Development: Phylogenetic p-value methods
We have developed methods to statistically score multiple sequence alignments for changes in evolutionary rates of substitutions. This will be used in the iSEEM project for studies of protein family evolution. These scores, called phylogenetic p-values, are negative log p-values from several tests for acceleration and/or deceleration of substitution rate. Test statistics include likelihood ratio tests, score tests, and functions of the expected numbers of substitutions (two published methods: SPH and GERP). These tests can be performed on the whole tree or a subtree (i.e. clade) of interest. The subtree tests allow detection of lineage-specific selection, enabling us to identify sites within proteins that may have experienced adaptive changes in a subset of species or in the ancestor of a group of living species.
Phylogenetic p-value calculations are performed on single alignment columns, making these some of the first molecular evolutionary methods that score evolutionary rates at single base pair resolution (the first to do so for nucleotide data). Scores from single sites can be combined to score genomic elements (e.g. alignment blocks, protein domains, or whole proteins). Single site scoring will be particularly useful in metagenomics applications, where many reads span only a small segment of the protein alignment. Since it will be difficult to assemble reads across the whole proteins, we may only have sequence data for a particular taxon at a few sites. With single-site scoring, we will be able to maximize our ability to conduct evolutionary analyses in the presence of so much missing data.
Development: Implementation
These methods have been implemented in a program called phyloP that is part of a freely available software package called PHAST available at http://compgen.bscb.cornell.edu/phast/. This project is joint work with Adam Siepel at Cornell University. In a separate project with Adam and colleagues at UCSF, we are applying these methods to study evolution in vertebrates. For the iSEEM project, Samantha Riesenfeld will be leading further extensions and evaluation of these methods, and their application to metagenomic data sets (e.g. Delong depth series and GOS) (Aim 2.1b).
Evaluation: Simulations
We have conducted extensive simulations to evaluate and compare these phylogenetic p-value methods. We have simulated DNA sequence alignments in two ways: (1) parametric and (2) non-parametric. Parametric simulations involve generating alignments from a collection of different (time-reversible, Markov) models of DNA evolution with either constant rates of evolution across a phylogenetic tree or rates that increase (or decrease) in a subtree of varying size. An advantage of these models is that we know precisely what processes are generating the observed simulation data. Non-parametric simulations involve sampling alignment columns from putatively neutrally evolving sequences (e.g. 4-fold degenerate sites) and conserved sequences (e.g. non-degenerate sites at second codon positions) from protein sequence alignments. Because they are generated from real data, these simulated alignments are more realistic (e.g. contain gaps, have more variable evolutionary rates). However, the disadvantage is that we do not know exactly how conserved the conserved sites are relative to the neutral sites, and this level of conservation is likely not constant across all conserved (second codon) sites.
For both types of simulation method, we have run phyloP to score evolutionary constraint at (i) single base pairs, (ii) short elements (5-25bp, representing subsegments of protein alignments in which we might have a consistent set of species and not too many gaps in a typical alignment of reads to a protein family), and (iii) longer alignments (50-200bp, representing more complete alignments of whole reads). We found that when alignments are deep enough (i.e. contain enough species) we can detect subtle changes in evolutionary rates at single base pair resolution (See figure below). Interestingly, the different methods have very similar performance. The SCORE method is appealing because it is faster than the other methods. The LRT has the advantage that it is a widely accepted approach. We will use one of these two methods in future work.
Performance of Four Phylogenetic P-Value Methods. We scored conservation at single sites (1bp) in a phylogeny of 40 species using parametrically simulated data with a typical level of conservation. This plot shows the rate at which false negatives (i.e. missed conservation) occur across the range of rates for false positives (i.e. incorrectly identified conservation). These are called ROC (Receiver Operating Curves). AUC is the area under the ROC curve. AUC=1 if no errors are made, and AUC=0.5 if the method is no better than random. False negative rates are low (also called high power) for all four methods. The times given (in seconds) are running times for the four methods. Running time is comparable (and acceptable for genome-scale studies) between LRT, GERP, and SPH tests. SCORE tests, however, are appreciatively faster. Performance is similar, though with slightly lower power, in a parallel simulation study of accelerated evolution (also on 1 bp alignments). As alignments get longer, AUC approaches 1 very quickly so that power is quite high for elements of 3-10bp. Lineage-specific tests have lower, but still reasonable, power.
To Do
There are several extensions of these methods we plan to implement in the next six months:
- extend methods to codon (or possibly amino acid) models
- evaluation of methods on simulated data that explicitly includes metagenomic reads
2.1b. Focused assessment of global evolutionary patterns and trends.
- Target date for completion 12/2009.
- Status: 5% complete. Expected completion 12/2009.
After developing a pipeline to do alignment and tree-building for protein families with metagenomic reads, we will process several metagenomic data sets, including the Delong depth series (a well controlled study of variable ecology at one geographic location) and the GOS (a global survey). This processing is already underway in the Eisen lab, with feedback from the Pollard and Green labs. Using protein alignments and trees for our collection of universal, even copy-number marker genes (Aim 1.1) we will first investigate variation in rates and patterns of evolution in these well-conserved genes using methods developed in Aim 2.1a and publicly available tools. Then, we will study correlations between this rate variation in biogeography and ecology of the sampling sites (Aim 3.1c). With this background for evolutionary patterns established, we will move on to similar studies using non-marker genes. Our goal will be to identify genes that are evolving uniquely (Aim 2.1c) (e.g. rapid subsitution rates, higher recombination, more gene gain or loss) in particular environments. We have done some initial thinking and planning for this project.
2.1c. Detailed investigation of particular cases of very rapid genome evolution
- Target date for completion 11/2010.
- Status: expected completion 11/2010.
Based on discoveries from Aim 2.1b, we will use bioinformatic and molecular evolutionary methods to investigate particular examples of gene families with unique patterns of evolution. Our goal will be to develop hypotheses about genetic adaptations and key innovations in the microbial tree of life.
2.1d. Integrate methods into CAMERA
- Target date for completion 11/2010.
- Status: expected completion 11/2010.
We will work with CAMERA to integrate our pipeline (including simulation, alignment, tree-building, and molecular evolutionary methods) into the CAMERA tool base.
2.2. Population Genomics
2.2a. Develop FST like measures of genomic variation within and between communities
- Target date for completion 12/2008.
- Status: in progress, essentially complete.
Steven Kembel and Jonathan Eisen have determined that FST statistics already implemented by Kembel in software he has developed (e.g., phylocom and picante) can be extended relatively easily to metagenomic data. We are working on adapting this methodology for metagenomics and expect to be completed shortly.
2.2b. Develop methods to quantify indels, recombination and rearrangement in comparison to reference genomes
- Target date for completion 12/2009.
- Status: expected completion 12/2009.
2.2c. Develop a genomic x spatial species concept for microbes
- Target date for completion 12/2009.
- Status: expected completion 12/2009.
2.2d. Develop phylogenetic sliding window method for binning
- Target date for completion 12/2009.
- Status: expected completion 12/2009.
2.2e. New measures and vulnerability of communities
- Target date for completion 11/2010.
- Status: expected completion 11/2010.
2.2f. Correlation of evolvability and community properties
- Target date for completion 11/2010.
- Status: expected completion 11/2010
3.1. Statistical metagenomics
3.1a. Survey CAMERA metagenomic data to determine scope of data types
- Target date for completion 12/2008.
- Status: Complete.
Joshua Ladau (with help from Samantha Riesenfeld)
Methods
A preliminary step of the iSEEM project is to use CAMERA to assemble information about existing metagenomic data. We compiled information on the types of metadata that are available for each project on CAMERA by consulting (i) individual metadata files for each project (on each project's webpage) and (ii) the listing of metadata on the CAMERA Project Samples webpage. We also gathered information on sequence data for each project by consulting files available for download on each project's webpage, published papers for each project, and the File Server Download page.
Results
Our findings are summarized in the Metadata and Sequencing Tables below.
- Click on a table to see a larger version:
The metadata available in the individual project files and those compiled on the Project Samples page are generally alike, although in a few cases, metadata are available in only one location. The available types of metadata fall into two broad categories: attributes of samples and attributes of locations from which samples were collected. In the former category, most projects provide date, location, and sample size information. In some cases, the projects also include information on sampling procedure used (e.g., filter size), number of organism sampled (e.g., bacteria abundance), and volume of substrate sampled. With regard to the location attributes, almost all studies provide information on the temperature, habitat type, and depth of samples. Other attributes of the abiotic and biotic environment are also often included, including major ion concentrations and biomass estimates. See the Metadata Table above.
With respect to the sequence data, all projects, except the Ocean Viruses and Moore Microbial Sequencing projects, make available raw reads, amino acid sequences, open reading frames, and ribosomal sequences. Six projects also provide assemblies. See the Sequencing Table above.
Conclusions and Recommendations
Overall, our experience working with CAMERA was positive. In general, the website is clearly designed, reliable, and fast. However, we did identify some aspects in which the website could be improved. First, the File Server Download page could be better organized, with data sets perhaps grouped by project or data type (e.g., nucleotide sequences, amino acid sequences, etc.). Given the size of many of the files, it would also be very helpful to have direct access to the database or command line access to the directories that contain the flat files. Second, while many files are accompanied by helpful documentation, additional documentation would at times be useful. For instance, it is unclear whether the ORF files available for different projects are equivalent.
We believe that the metadata and sequence data available on CAMERA will be useful in the development and application of new approaches to metagenomic data analysis. Cataloguing the available data comprises an important initial step towards reaching these goals.
3.1b. Methods for model selection in metagenomics
- Target date for completion 12/2009.
- Status: 5% complete; expected completion 12/2009.
Using simulations and real data, we will evaluate existing statistical approaches to model selection for their utility for assessing correlations between genome sequence data (and summaries thereof, such as diversity or molecular evolutionary statistics) and biogeographic/ecological metadata. We have started thinking about this problem and planning our approach.
3.1c. Correlation analysis
- Target date for completion 12/2009.
- Status: 15% complete; expected completion 12/2009.
Using the models developed in Aim 3.1b, we will develop methods for estimation and testing of correlations between sequence and metadata. We have done some initial planning for this subaim. A rencetly published methodological paper by Katie Pollard and Mark van der Laan (UC Berkeley) that establishes the statistical concept of a supervised distance matrix will be useful here. This paper was motivated by gene expression data, rather than sequencing data. And the metadata in the motivating problem was clinical outcome data, such as patient survival. However, the general framework, which allows one to cluster genes with respect to their correlation with a continuous metadata variable could be easily extend and applied to clustering sequence reads or rates of evolution therein with respect to their correlation to biogeographic metadata.
3.1d. Apply correlation analysis to public data
- Target date for completion 12/2009.
- Status: expected completion 12/2009.
We will apply the methods developed in Aims 3.1b and 3.1c to conduct correlation analyses using global metagenomic data sets.
3.1e. Integrate into CAMERA
- Target date for completion 11/2010.
- Status: 5% complete; expected completion 11/2010.
We will work with CAMERA to integrate our correlation tests into the CAMERA tool base.
C. Communications, Collaboration, Outreach and Education
Project group page at Open Wet Ware
Initially, we used a collaborative Wiki site hosted by the Eisen lab as a place to post and share information between labs. This became a bit cumbersome to host since we are not experts in Wiki software development. After exploring many options we have chosen to use Open Wet Ware (http://openwetware.org/wiki/) because it is part of an "Open Science" initiative that is consistent with the open goals of the iSEEM project. In addition, the Open Wet Ware developers have implemented a "private" page option where researchers can share information that one does not want to make public. We now use this site for sharing, note taking, and collaborative discussions. In addition we are developing a "Public" page for the project at http://openwetware.org/wiki/ISEEM.
CiteYouLike
We are keeping a bibliography of relevant literature on CiteYouLike (http://www.citeulike.org/groupfunc/6072). This is a publicly available collection (although only group members can edit it).
Collaborations
- Katie Pollard is collaborating with Adam Siepel (Cornell University) to develop, test and implement molecular evolutionary methods for identifying gene families with unusually fast or slow rates of evolution.
- The Pollard lab has been discussing alignment and tree-building with Steven Brenner (UC Berkeley) and his colleagues. Jonathan Eisen will be joining these discussions.
- Steven Kembel is collaborating with the NCEAS ecophylogenetics working group to develop phylogenetic diversity metrics that can be applied to metagenomic data sets
- James O'Dwyer is collaborating with Helene Morlon (University of Oregon) to generate predictions for phylogenetic beta-diversity using coalescent theory.
- Jessica Green has been discussing the potential for exploring microbial trait-based biogeography using metagenomic data with Brendan Bohannan at the University of Oregon. Green and Brendan recently received funding from the National Center for Ecological Analysis and Synthesis NCEAS to organize a working group on this subject. Eisen and Pollard have been invited to participate in this working group.
- The Green lab is collaborating with Steve Giovannoni (Oregon State University), Alexandra Worden (Monterey Bay Aquarium Research Institute), and Craig Carlson (UC Santa Barbara) on a GBMF funded pryrosequencing project to explore microbial diversity along a depth gradient at the Bermuda Atlantic Time-series Study (BATS).
Courses
- Jonathan Eisen taught a seminar in Winter 2007 students in the Population Biology Graduate project on metagenomics and phylogenomics.
- Jonathan Eisen taught a course in Spring 2008 on "Microbial phylogenomics" where ~ half of the course focused on microbial diversity.
- Jonathan Eisen taught a lecture for the Bodega Bay Workshop in Applied Phylogenetics in March 2008 (see http://bodegaphylo.wikispot.org/Front_Page for more detail). A component of the lecture focused on microbial diversity and metagenomics.
- Jonathan Eisen is designing a metagenomics course for Spring 2009 at UC Davis. All course material will be made available online. In addition it will be audio and possibly video recorded and made available through iTunes.
- We are exploring the possibility of jointly offering a metagenomics class for the community at Bodega Bay in the Summer of 2009.
Presentations / Conferences
- Jessica Green presented iSEEM research at the 2008 Gordon Conference Metabolic Basis of Ecology (Biddeford; July 2008). She is currently scheduled to present at the 2009 Gordon Research Conference Applied and Environmental Microbiology (South Hadley; July 2009) and an organized symposium on the human microbiome Ecological Society of America Ecological Society of America Annual Meeting (Albuquerque; August 2009).
- Steven Kembel will present iSEEM research at the University of Michigan Young Scientists Symposium (Ann Arbor; March 2009), an organized symposium on phylogenetic ecology at the Ecological Society of America annual meeting (Albuquerque; August 2009), and a bioinformatics workshop for FESIN members at Botanical/Mycological Society of America annual meeting (Snowbird; July 2009).
- James O'Dwyer presented iSEEM research at the 2008 Gordon Conference Metabolic Basis of Ecology (Biddeford, ME; July 2008), at ESA 2008 [3] (Milwaukee, WI; August 2008), and at The 16th International Microbial Genome Conference. Sept 14-18, 2008 Lake Arrowhead, CA [4] (Lake Arrowhead, CA; September 2008).
- Martin Wu and Jonathan Eisen presented a posted "A Simple, Fast and Accurate Method of Phylogenomic Inference" at the The 16th International Microbial Genome Conference. Sept 14-18, 2008 Lake Arrowhead, CA
- Jonathan Eisen gave presentations including some of the results from this project at the International Metagenomics Meeting in November 2008, the ASM General Meeting in May of 2008 (video of the talk is available here http://video-jsoe.ucsd.edu/asx/Metagenomics08/WednesdayNovember5/Eisen.asx), and the AGBT meeting in Marco Island in February 2008.
Publications
- Wu M, Eisen JA. A Simple, Fast and Accurate Method of Phylogenomic Inference. Genome Biology 9: R151. Paper available fully "open access" at http://genomebiology.com/2008/9/10/R151.
- Software available freely at http://bobcat.genomecenter.ucdavis.edu/AMPHORA/.
- Evaluated by Faculty of 1000 (see http://www.f1000biology.com/article/id/1134841).
- Wu D, Hartman AL, Ward N, Eisen JA. PLoS ONE 3(7): e2566. An Automated Phylogenetic Tree-Based Small Subunit rRNA Taxonomy and Alignment Pipeline (STAP). Paper available fully "open access" at http://www.plosone.org/article/info:doi/10.1371/journal.pone.0002566.
- Software available freely at http://bobcat.genomecenter.ucdavis.edu/mediawiki/index.php/STAP_download
- Pollard, K.S., van der Laan, M.J. 2008. Supervised distance matrices. Statistical Applications in Genetics and Molecular Biology 7(1): article 33. See http://www.bepress.com/sagmb/vol7/iss1/art33/ and File:SDM.pdf.
- Green, J., Bohannan, B., Whitaker, R. 2008. Microbial biogeography: from taxonomy to traits. Science 320: 1039- 1042. See http://www.sciencemag.org/cgi/content/full/320/5879/1039 and File:Green et al. Science 2008.pdf.
- Bryant, J.B., Lamanna, C., Morlon, H., Kerkhoff, A.J., Enquist, B.J., Green, J.L. 2008. Microbes on mountainsides: Contrasting elevational patterns of bacterial and plant diversity. Proceedings of the National Academy of Sciences 105 Supplement 1: 1505-11511. See http://www.pnas.org/content/105/suppl.1/11505 and File:Bryant et al. PNAS 2008.pdf.
- O'Dwyer, J.P., Lake, J.L., Ostling, A., Savage, V.M., Green, J.L., An integrative framework for stochastic, size-structured community assembly. In press at Proceedings of the National Academy of Sciences. See File:Stochastic size manuscript revised3.pdf.
- A manuscript on molecular evolutionary methods (by Katie Pollard) is in preparation.
II. Group Meetings
Bi-weekly PI-only conference calls
The three PIs meet bi-weekly by phone or skype conference call. These calls focused mainly on logistical issues (e.g. hiring, website, computing, reports) and strategic planning (e.g. collaborations between labs, shared resources). Notes from all calls are available on our wiki at http://iseem.private.openwetware.org/wiki/ISEEM-Conference-calls.
Bi-weekly full group conference calls
The full group (PIs, postdocs, collaborating lab members) meet bi-weekly by skype conference call. These calls focus mainly on scientific issues, in particular projects that are collaborative or impact the aims of different labs (e.g. alignments and trees for marker genes, simulation methods, diversity measures). Notes from all calls are available on our wiki at http://iseem.private.openwetware.org/wiki/ISEEM-Conference-calls.
Monthly get-togethers
The postdocs and graduate students of the Eisen and Pollard labs have initiated regular visits (every three to four weeks) to encourage an informal exchange of scientific questions, ideas, and techniques related to the iSEEM project. Given the variation in scientific background among the collaborators, these face-to-face discussions are especially helpful in cohering our research efforts. The first get-together took place in early January at Gladstone Institutes (at UCSF); the next one is scheduled for the end of January at UC Davis. We plan to use skype sessions to include the postdocs and graduate students from the Green lab in our meetings. Green lab members may attend in person occasionally.
Other meetings
- Katie Pollard, Joshua Ladau, Jonathan Eisen, and Jonathan Eisen's lab group met at UC Davis on September 22. Research directions for Josh were discussed, including development of optimal community phylogenetic statistical methods, and methods for inferring species richness and abundance from metagenomic data. Josh will be starting as a postdoc in Katie Pollard's laboratory on October 22.
- Jonathan Eisen, Martin Wu, James O'Dwyer, and Liz Perry held a meeting at the Lake Arrowhead small genomes conference to discuss methods for calculating metagenomic distances.
Quarterly meetings
April 2008
We held a quarterly meeting April 18, 2008 at CalIT2 on the UCSD campus as part of a meeting with the CAMERA team. The people from the iSEEM project at the meeting were: Jonathan Eisen, Martin Wu, Dongying Wu, Jessica Green, James O' Dwyer, Liz Perry, Katherine Pollard. Multiple personnel from the CAMERA team were also there. The meeting consisted of a discussion of the goals CAMERA as well as the goals of the iSEEM project as well as the CAMERA subcontract to the Eisen lab. We then discussed how the iSEEM team could work with CAMERA both to get the science done that is part of the iSEEM project as well as to implement in CAMERA any tools the iSEEM project develops. Overall the meeting was very helpful. It did however highlight some challenges with working with CAMERA to get exactly what the iSEEM project needed in terms of scientific resources. Follow up discussions with people from CAMERA including Paul Gilna and Mark Ellisman have led to a strategy to move forward that seems likely to solve all of the perceived challenges.
September 2008
We had a joint group meeting with people from CAMERA, the Kepler Workflow Project, and our group. The meeting was held at UC Davis in the Genome Center and involved the following people
- UCSD/CAMERA: Amarnath Gupta, Ilkay Altintas, Jeffrey S. Grethe, Paul Gilna
- Kepler-Davis: Bertram Ludascher, Shawn Bowers, Timothy McPhillips, Sean Riddle
- Eisen Lab/iSEEM: Amber Hartman, Jonathan Eisen, Martin Wu, Srijak Bhatnagar, Dongying Wu
In the meeting we discussed how to work with CAMERA to take methods developed from the iSEEM project and integrate them as Kepler Workflows (http://kepler-project.org/) within the CAMERA system. We came up with a plan for the next few months of work which involves first working on a rRNA analysis workflow and using it as a test to see how to take workflows from the iSEEM project and develop them into CAMERA tools. Once this is done we will move on to protein analysis workflows.
December 2008
We held a Quarterly meeting in December of 2008 over two days at U. C. Davis. Participants were Jessica Green, Steve Kembel, James O'Dwyer, Katie Pollard, Josh Ladau, Samantha Riesenfeld, Jonathan Eisen, Martin Wu, Dongying Wu, Sourav Chatterji, Aaron Darling, Kelly Kryc, Srijak Bhatnagar. All researchers gave short presentations of their work. In addition we discussed multiple topics including metagenomics education, outreach, simulations, scientific workflows, data types, phylogenetic reconstruction, community diversity, and measuring selection. The PIs held a separate meeting with Kelly Kryc to discuss the annual report and other programmatic issues.
III. Unexpected Challenges
Hiring
Hiring took a bit longer than we predicted. The team was fully assembled by our Quarterly Meeting at UC Davis in December 2008. Most postdocs started in Fall 2008, rather than Summer 2008. Several projects due in 2008 are therefore less than 100% complete.
Working with CAMERA
A second minor challenge has come up in terms of figuring out how to take products (e.g., software, algorithms, data sets) we develop and have them integrated into CAMERA. Initially, our interactions with CAMERA were pleasant but not very productive as nobody there was able to help us develop any plan for how we would work with them. Recently, we have had more success with discussions with CAMERA staff about how to move forward. We are working with CAMERA staff to develop a plan for doing this but as of yet we have not had significant success in achieving any results. We believe it may be necessary to have a more hands on involvement from the Gordon and Betty Moore Foundation to help guarantee that the results of our work end up being made available to the community through CAMERA. We note that ALL of our tools and data sets that we develop will be made available for free to the entire research community through sites like Sourceforge and the iSEEM project page. However, it is our goal to work with CAMERA to make our products available not just as separate pieces but as an integrated whole within the CAMERA portal.
IV. Financial reporting
For the original Year 1 budget $332,205 was allocated to expenses at UC Davis and $217,100 for subcontracts to U. Oregon and UCSF.
As of January 30, 2009, we estimate that ~$130,000 remains in the Davis allocation and ~$30,000 in the subcontract allocation (this estimate includes as "spent" an order that has been placed for a computational cluster for $30,000 as well as invoices from U. Oregon and UCSF that are being processed). An excel spreadsheet is attached with more detail on these numbers.
In total this means we have underspent the Year 1 budget by about 29%. Most of this underspending was due to the delays in hiring personnel for the project as discussed in earlier reports and summarized in this report. In addition, the move of Dr. Pollard to UCSF created some delays in spending.
In order to achieve all of the outcomes for the project as originally planned we need to hire 1-2 additional personnel to make up for the missed effort due to the delays in hiring and/or when personnel started work. We have offered a job to one PostDoc who will work jointly between the Pollard and Eisen labs and are interviewing another candidate for the Eisen lab this week. In addition, we are working on a subcontract for Martin Wu who has left the Eisen lab for a faculty position at U. Virginia. Together with these new hires and the subcontract, we project we will expend our full Year 2 allocation and the leftover funds from Year 1 by the end of Year 2.
We estimate that it will take us approximately 1.5-2 additional months to spend down the remaining funds from Year 1 and thus will not need the Year 2 allocation until ~March 15,2009. We note that this is an estimate based upon trends in current spending.