IGEM:IMPERIAL/2009/M2/Modelling/Analysis
<html> <style type="text/css"> /*----------------------------------------------------------------------------*/ /* Structure */ /*----------------------------------------------------------------------------*/
- II09_mainWrapper{
width: 800px; align: center; border: none; margin: 0px auto; padding: 0px;
}
/*----------------------------------------------------------------------------*/ /* Content */ /*----------------------------------------------------------------------------*/
- II09_content_wrapper {
width: 550px; float: left; margin-top: 5px; margin-bottom: 5px; position:relative;
}
- II09_content_boxtop {
width: 550px; height: 10px; background:url(http://2009.igem.org/wiki/images/c/c8/II09_content_top.png) no-repeat bottom left; position: relative; float: left;
}
- II09_content_boxbottom {
width: 550px; height: 10px; background:url(http://2009.igem.org/wiki/images/b/b4/II09_content_bottom.png) no-repeat top left; position: relative; float: left;
}
- II09_content {
width: 550px; padding: 10px; background-color:#ffffff; border-left: 1px solid #ccc; border-right: 1px solid #ccc; position: relative; float: left; font-size: 9pt; text-align: justify;
}
/*----------------------------------------------------------------------------*/ /* Tabs */ /*----------------------------------------------------------------------------*/
.II09_Tabs_Blue {
float:left;
width:100%;
background:#CCCCFF url("http://openwetware.org/images/b/bb/SDbg.gif") repeat-x bottom;
font-size:93%;
line-height:normal;
margin: 0;
padding: 0;
}
.II09_Tabs_Blue ul {
margin:0; padding:10px 10px 0; list-style:none;
} .II09_Tabs_Blue li {
float:left;
background:url("http://openwetware.org/images/b/b7/SDleft.gif") no-repeat left top;
margin:0;
padding:0 0 0 9px;
}
.II09_Tabs_Blue b {
float:left;
display:block;
background:url("http://openwetware.org/images/f/f8/SDright.gif") no-repeat right top;
padding:5px 15px 4px 6px;
text-decoration:none;
font-weight:bold;
color:#765;
} </style> </html>
<html> <body> </html> <html>
<head> <style type="text/css">
- II09_Header{
border-top: 1px solid #7b7b7b; border-left: 1px solid #7b7b7b; border-right: 1px solid #7b7b7b; border-bottom: none;
background: url(http://2009.igem.org/wiki/images/3/39/II09_Banner.jpg); background-position: right -75px; width: 783px; height: 150px; margin-left:15px; align: right;
} </style> </head>
<body>
</body> </html> <html> <head> <style type="text/css">
- II09_Breadcrumb{
backgrounewd: #7b7b7b; background: url(http://2008.igem.org/wiki/images/c/c8/Tudelft_menu_header_bg_1.gif); border-top: none; border-left: 1px #7b7b7b solid; border-right: 1px #7b7b7b solid; border-bottom: 1px #7b7b7b solid; filter:alpha(opacity=100); -moz-opacity:1.00; opacity:1.0; text-color: #000000; padding-left: 10px; margin-left: 15px; width: 773px;
} </style> </head>
<body>
<script type="text/javascript">
var address = document.location.href; var addressSplit = address.split("/");
document.write('<a href="http://2009.igem.org/' +addressSplit[3]+ '">Home (' +addressSplit[3]+ ')</a>');
for (i=4;i<8;i++){
if (addressSplit[i]!=undefined){
document.write(' | ');
document.write('<a href="' +addressSplit[i]+ '">' +addressSplit[i]+ '</a>');
}
}
document.write('');
</script>
</body> </html> <html> <style type="text/css"> /*----------------------------------------------------------------------------*/ /* Menu */ /*----------------------------------------------------------------------------*/
- menu {
width: 200px; margin: 5px 15px 0 15px; position: relative; float: left;
}
- menu ul {
padding: 0; margin: 0 0 10px 0; list-style: none;
}
- menu li {
margin:0;
}
- menu a {
display: block; width: 185px; height: 19px; margin-top: 1px; margin-left:15px; background: url(http://openwetware.org/images/4/46/II09_Menu_ItemHover.png) no-repeat right top; padding: 2px 4px; text-decoration: none; color: #fff;
}
- menu a:hover {
display: block; width: 185px; height: 19px; margin-top: 1px; margin-left:15px; background: url(http://openwetware.org/images/b/bf/II09_Menu_Item.png) no-repeat right top; padding: 2px 4px; color: #000;
}
- menu .selflink {
/*font-weight:bold; width: 200px; border: 1px solid #bbb; background: url(http://2008.igem.org/wiki/images/c/c8/Tudelft_menu_header_bg_1.gif) repeat-x bottom; color: #111; padding: 2px 4px;*/
display: block; width: 200px; height: 30px; margin-top: 1px; border: 1px solid #bbb; background: url(http://2008.igem.org/wiki/images/c/c8/Tudelft_menu_header_bg_1.gif) repeat-x left bottom; padding: 2px 4px; color: #000;
}
- menu a.external {
margin: 0; padding: 0; width: 156px;
}
.menu_header {
font-weight:bold; width: 200px; border: 1px solid #bbb; background: url(http://2008.igem.org/wiki/images/c/c8/Tudelft_menu_header_bg_1.gif) repeat-x bottom; color: #111; padding: 2px 4px; text-align:left;
} </style>
</html> <html>
<img src="http://2008.igem.org/wiki/images/a/a1/Tudelft_content_top.png" width="572">
<script type="text/javascript">
var gaJsHost = (("https:" == document.location.protocol) ? "https://ssl." : "http://www.");
document.write(unescape("%3Cscript src='" + gaJsHost + "google-analytics.com/ga.js' type='text/javascript'%3E%3C/script%3E"));
</script>
<script type="text/javascript">
try {
var pageTracker = _gat._getTracker("UA-11206997-1");
pageTracker._trackPageview();
} catch(err) {}</script>
</html>
Preliminary Model 1: Genetic circuit
Hypothesis
- When glucose concentrations are above threshold, there would not be any activation of production of colanic acid or trehalose.
- When glucose concentrations decrease below threshold, there will be strong activation of M2 and there will be an increase in protein production in the system.
- For decreased concentration of glucose, there will be increased amounts of colanic acid and trehalose produced.
Equations
[math]\displaystyle{
\begin{alignat}{2}
\frac{d[m_i]}{dt} = \frac{km_i^{n_i}}{{[g]}^{n_i}+ km_i^{n_i}} - dm_i*[m_i] \qquad (1)\\
\frac{d[p_i]}{dt} = [m_i]*kp_i - dp_i*[p_i] \qquad (2)\\
where\ i = 1, 2, 3, 4, 5
\end{alignat}
}[/math]
Legend
| Term | Variable/Parameter | Meaning/description |
| 1 | - | RcsB |
| 2 | - | B3023 |
| 3 | - | Waal Ligase |
| 4 | - | OtsA |
| 5 | - | OtsB |
| g | Variable | Concentration of glucose |
| mi | Variable | Concentration of ith mRNA |
| pi | Variable | Concentration of ith protein |
| ni | Parameter | Hill coefficient of ith mRNA |
Explanation of equations
- In equation (1), we have used a repressing hill function of glucose. This is because glucose, represses CRP, which activates our system. A decrease in [glucose] below threshold will activate M2 strongly.
- The genes are produced in parallel, hence the repressing hill function will appear in the mRNA equations, for all our proteins.
Simulations
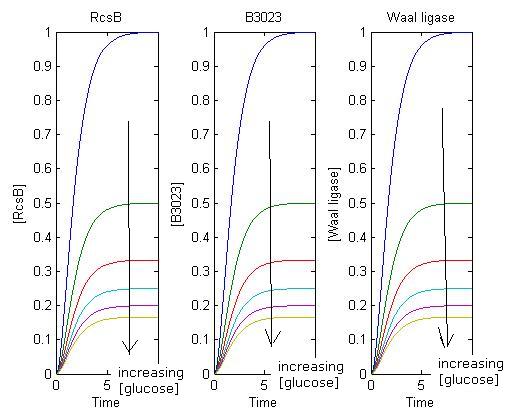
Simulation 1: Effects of increasing glucose concentrations on production of protein
Explanations
- When there is a decrease in glucose concentration beyond threshold, there is a great increase in amount and rate of protein production.
- A short time delay is seen as transcription and translation takes time.
Data Fitting
References
<html>
<img src="http://2008.igem.org/wiki/images/8/89/Tudelft_content_bottom.png" width="572">
</body> </html>
<html> <style type="text/css">
- II09_Footer{
width:763px; margin-left: 15px; background: url(http://2008.igem.org/wiki/images/c/c8/Tudelft_menu_header_bg_1.gif) repeat-x; background-position: bottom; padding: 10px; border-top: 1px #7b7b7b solid; border-left: 1px #7b7b7b solid; border-right: 1px #7b7b7b solid; border-bottom: 1px #7b7b7b solid; position:relative; float: left;
} </style>
<body>
</body> </html>