IGEM:IMPERIAL/2009/M2
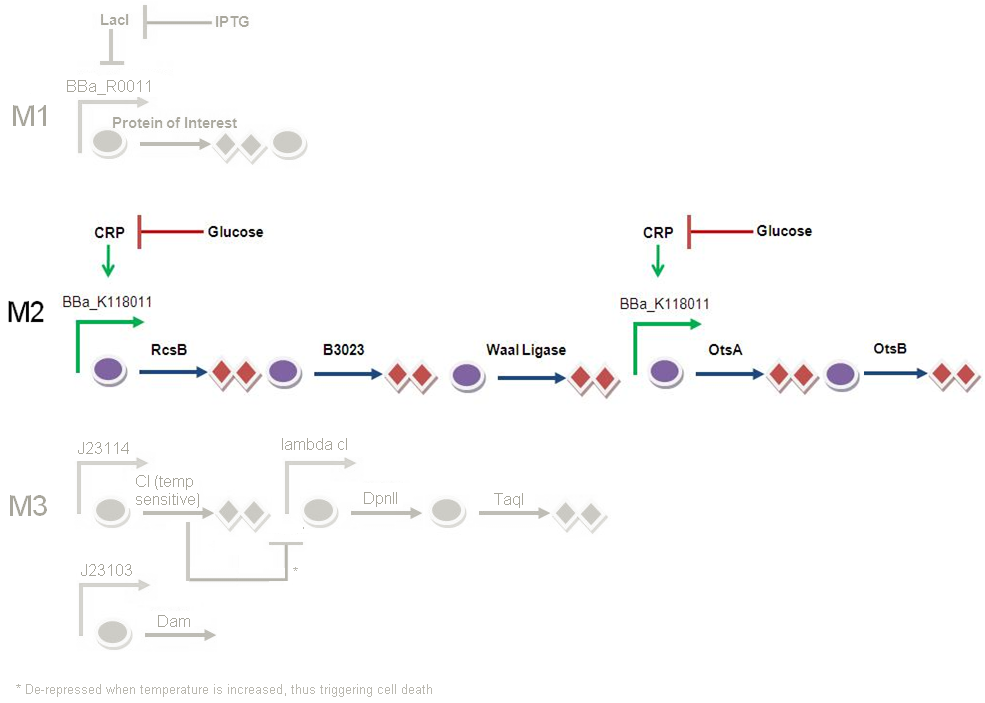
Module 2 - Genetic Circuit
Glucose is originally present in our media, and it is repressing CRP, and in turn M2 will not be activated. When the e. coli cells consume glucose in the media, there will be a decrease in glucose concentration. This decrease in glucose concentration will result in a decrease in repression on CRP. CRP will then activate BBa_K118011 and kick start production of colanic acid (controlled by RcsB, B3023 and waal ligase) and trehalose (controlled by OtsA and OtsB). This will mark the beginning of M2.
Module 2 - Encapsulation
- People: James, Nuri and Dave
Module 2 consists of three sub-sections that require characterisation: 1) Promoter (Nuri)
2) Colanic acid (James) Colanic acid is a negatively charged polymer that protects the the E.coli cell from environmental stresses - namely low pH. By up-regulating colanic acid biosynthesis we hope to augment protection against low pH.
3) Trehalose (Dave)
Colanic acid
Can the chassis produce colanic acid?
COMPLETE
Background:
While there are multiple strains of E.coli K12 that can produce colanic acid, it has been noted in the literature that some cannot. It is important that before selecting our K12 chassis, we ensure that it can produce colanic acid. A procedure for this has been prepared in Assay 2.1
Data Analysis:
N/A, - Data is qualitative
Protocol:
http://openwetware.org/index.php?title=IGEM:IMPERIAL/2009/M2/Assays/2.1
Influence of carbon source on colanic acid production & population growth
COMPLETE
Background: Once the glucose in the media has been consumed, the chassis will utilise a secondary carbon source to faciliate the production of colanic acid. Assay 2.2 serves to determine the secondary carbon source that is most suited to colanic acid production.
Data Analysis:
This assay will produce the following data types:
Optical density = OD (600nm)
Packed Cell Volume (PCV)
Average amount of colanic acid produced per cell can be calculated in the following way:
1) Measure PCV and OD of colanic acid producing cells.
2) Divide the PCV by the OD to facilitate the relative quantification of the amount of colanic acid produced per cell.
3) Repeat this for different carbon sources to determine which carbon source is most suited to colanic acid production.
Protocol:
http://openwetware.org/index.php?title=IGEM:IMPERIAL/2009/M2/Assays/2.2
Sodium Acetate Encapsulation
POSTPONED
Background:
The encapsulation of colanic acid coated E.coli with sodium acetate has never been attempted. Theoretically, the highly negativly charged colanic acid coat should act as a nucleation site for Na+ ions in the sodium aceteate solution, but it would be wise to prove this in the lab as quickly as possible.
Since we can induce colanic acid production chemically, Assay 2.3 will provide a quick answer as to whether we need to investigate alternative secondary encapsulation technologies.
Data Analysis:
Protocol:
http://openwetware.org/index.php?title=IGEM:IMPERIAL/2009/M2/Assays/2.3
Sodium Acetate Exothermic Crystalisation
POSTPONED
Background: Sodium acetate crystalisation is an exothermic reaction (hence the use of sodium acetate in hand warmers). If we want exothermic crystalisation to flip the kill switch, we need to model the heat that is released upon crystalisation.
Data Analysis:
Protocol:
http://openwetware.org/index.php?title=IGEM:IMPERIAL/2009/M2/Assays/2.4
Qualitative identification of construct-induced colanic acid production
COMPLETE
Background: This assay indicates the successful expression of colanic acid inducing genes. Successful transformation is indicated by visualisation of GFP while the ability of the construct to induce colanic acid biosynthesis is reflected by the appearance of a mucoid colony.
Construct 2.1:
BBa_F2620 - BBa_B0034 - BBa_K200000 - BBa_E0240
Construct 2.2:
BBa_F2620 - BBa_B0034 - BBa_K200000 - BBa_K200002 - BBa_E0240
Construct 2.3:
BBa_F2620 - BBa_B0034 -BBa_K200000 - BBa_K200002 - BBa_K200003 BBa_E0240
Data Analysis:
Protocol:
http://openwetware.org/index.php?title=IGEM:IMPERIAL/2009/M2/Assays/2.5
Quantitative analysis of construct-induced colanic acid production
COMPLETE - Seek advice on cloning strategy...
Background: This assay faciliates an assessment of the protective effect of differing colanic acid profiles.
Various combinations of colanic acid stimulating genes will produce different colanic acid profiles. Profiles are predicted to vary with respect to both amount of colanic acid produced and additionally the type (e.g. tethered to outer membrane or non-covalently attached). Each construct should be regarded as a different transfer function that converts a PoPs input to a colanic acid output.
Quantity of colanic acid can be determined by a packed cell volume (PCV) assay while quality can be determined by the number of cells that remain intact after exposure to simulated gastric conditions. This data can subsequently be used to determine the quality per volume of colanic acid produced. Ideally, the construct that provides the greatest protection with the smallest amount of colanic acid will be selected for use in our final design.
Genetic Constructs:
The following three constructs allow three different transfer functions to be independently assessed.
Construct 2.1:
BBa_F2620 - BBa_B0034 - BBa_K200000 - BBa_E0240
Construct 2.2:
BBa_F2620 - BBa_B0034 - BBa_K200000 - BBa_K200002 - BBa_E0240
Construct 2.3:
BBa_F2620 - BBa_B0034 -BBa_K200000 - BBa_K200002 - BBa_K200003 BBa_E0240
Protocol:
http://openwetware.org/index.php?title=IGEM:IMPERIAL/2009/M2/Assays/2.6
2) Trehalose
Using a characterised POPs input, we aim to relate this to a quantitative trehalose output. This can be done using a trehalose assay. As a functionality assay, we aim to relate trehalose prodution to cell stress resistance; for example, dessication or temperature resistance. As the function of the trehalose is to protect our protein of interest, we would like to quantify the change in resistance of proteins to conformational change to concentration of trehalose. We have chosen to link our trehalose concentration to resistance of a standard protein, catalase, the concentration of which can be measured using a standard hydrogen peroxide assay.
DNA construct
For trehalose testing constructs we only need one testing construct of both genes, which will produce both enzymes needed for trehalose production.
BBa_F2620 - BBa_B0034- OtsA - BBa_B0034 - OtsB -BBa_B0015
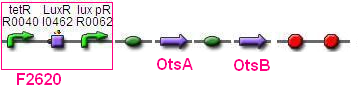 Assay both genes seperately??
Assay both genes seperately??
Cloning strategy (assembly)
- James Field 14:30, 11 August 2009 (EDT):Dave, can you put in the links for Escherichia coli BL21(DE3) sequences
- James Field 08:20, 12 August 2009 (EDT): Dave, I checked the links that you put in for OtsA and OtsB. They appear to be the ones for Escherichia coli K-12 substr. MG1655 and not Escherichia coli BL21(DE3). Can you correct this.
Blast search between Escherichia coli K-12 substr. MG1655 and Escherichia coli BL21(DE3).
BL21 (DE3) : Genome
| Gene | Strain | Score | Expect | Identities | Gaps | Link to sequence |
| OtsA | Escherichia coli BL21(DE3) | 2632 bits (1425) | 0.0 | 1425/1425 (100%) | 0/1425 (0%) | OtsA Link (Reverse Compliment) |
| OtsB | Escherichia coli BL21(DE3) | 1480 bits (801) | 0.0 | 801/801 (100%) | 0/801 (0%) | OtsB Link (Reverse Compliment) |
| RcsB | Escherichia coli str. K-12 substr. DH10B | 1203 bits (651) | 0.0 | 651/651 (100%) | 0/651 (0%) | DH10B Sequence |
| B3023 (ygiV) | Escherichia coli str. K-12 substr. DH10B | 893 bits (483) | 0.0 | 483/483 (100%) | 0/483 (0%) | DH10B Sequence |
| rfal (Waal) | Escherichia coli str. K-12 substr. DH10B | 2327 bits (1260) | 0.0 | 1260/1260 (100%) | 0/1260 (0%) | DH10B Sequence |
Please click the link above to reach the Assays page.
Output data
2) Colanic Acid
1) Determine the average colanic acid produced per cell for a given PoPs input.
Average colanic acid production can be determeind by three pieces of information: PCV (µl), OD at 600nm (pre-PCV) and HSL input (mM).
2) Determine the protective effect of the colanic acid for a given PoPs iput.
This can be determined by two pieces of information: OD (pre-low pH exposure) & OD (post-low pH exposure)
3) Determine colanic acid quality - e.g. do the same volumes of colanic acid offer different degrees of protection between constructs?
This can be determined using the data obtained in steps 1) and 2).
3) Trehalose
For a given transfer function:
1) Average trehalose produced per cell for agive POPs input. This can be found by getting the overall quantity of trehalose for a sample of the culture and normalising against the cell density given by the OD at 600nm.
2) Protection against protein conformational changes offered by trehalose. As we will not be expressing our enzymes of interest along with our trehalose/encapsulation module, we instead would choose to monitor the effects on a common protein found within e-coli. Catalase is such a protein, and assays can be performed to monitor changes in activity when exposed to environmental stresses.
Please click the title to go to the modelling directions for this module.