IGEM:IMPERIAL/2009/4Sep/System Overview/Mod3
Module 3
Module function in overall design
- High level description
This module serves to remove the genetic material from the bacteria after both protein production and encapsulation have been completed.
This serves to allay any genetically modified organism concerns, and is useful as a reusable module for other projects.
2 main parts:
1) Thermoinduction
2) Restriction Enzyme killing
- Input
Temperature increase to 42 degrees Celsius will trigger Module 3
- Output(s)
Restriction Enzyme DpnII and TaqI activity leads to cell death
Assays to be done
| Assays | Importance (1=Most important. 3=Least important) | When? (1=Now. 3=Last) | Variable | Rationale | Output | Model |
| Staining assay/CFU | 1 | - | Temperature | Effect of temperature on viable population | Fluorescence ratio/ CFU | Yes |
| Restriction enzyme assay | 3 | Now | Restriction enzyme concentration | "Visualise the effects of restriction enzymes on DNA.Can we do methylase?" | Gel | N/A |
| Harvard temperature | 2 | - | Temperature | How does temperature influence GFP output | Fluorescence | No |
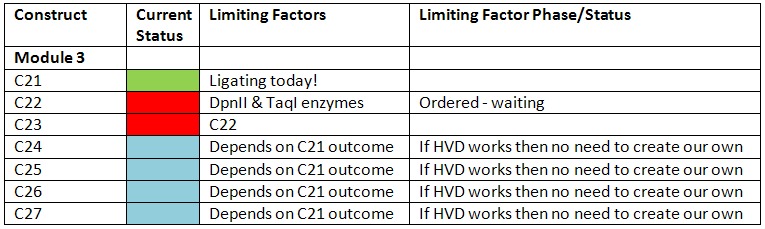
Testing Construct Design
- Total Number of testing constructs
Definite constructs: 2
Possible constructs: 3 (depending on if Harvard GFP works)
For all the constructs currently being designed - see here
- Rationale behind testing constructs
To test how temperature will affect either the GFP expression, or the efficiency of killing.
Progress
- Current status in terms of cloning for each assembly
- The Harvard-GFP is the main ligation, from which testing can start.
- For the second testing construct, we are awaiting DpnII and TaqI from MrGene
- To save time, the self-synthesized cI and pLambda will not be constructed until after Harvard-GFP has been tested
What can be completed in time for Jamboree
- Will definitely finish the Harvard-GFP testing, but the rest will depend on when Resriction enzymes come and ligation success
Data we hope to get from this
NO DNA construct
To test how the concentration of restriction enzymes and methylases affects the cutting of genomic DNA.
- will get a Yes/No answer for each set concentration of restriction enzymes and methylases.
Importance:
- in showing that the restriction enzymes work and that there is a balance with the methylases (the concentration can be experimentally determined)
Assay:
- Genomic preparation assay to test if it can be cut
How essential in whole project: 4
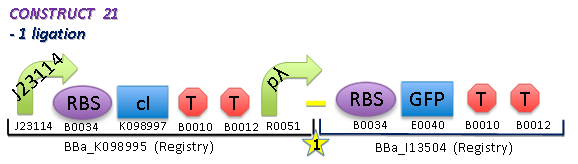
To test how the GFP fluorescence will vary with the temperature
- will get fluorescence output(A.U.) over time for 28 degrees and 42 degrees
In addition, since GFP is temp sensitive, need to also find out how GFP fluorescence varies with temperature
use
- test GFP fluorescence varying with temperature using any promoter with GFP eg CRP promoter
Importance:
- in showing that the thermoinduction system works- trigger for killing
Assay:
- Promoter characterisation assay and the effect of increasing temperature
- Essentially just monitering GFP fluorescence over time at different temperatures
How essential in whole project: 6
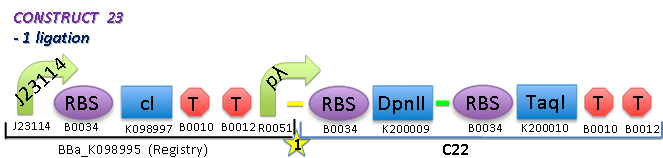
To test the effect of temperature on the killing efficiency of the restriction enzymes
Importance:
- In showing that the thermoinduction system and the killing strategy works- trigger for killing
Assay:
- Staining Assay which gives the percentage of viable and dead cells over time
- Important for modelling killing rate
How essential in whole project: 8
Ready for testing construct
- Protocols written and reviewed ?
GFP fluorescence and Harvard YES
Staining Assay YES
Restriction Enzyme and Genomic Assay YES
- Any expertise needed? Acces to specific equipment needed ?
Multiplate reader
- Modelling done ? Ready to analyse experimental data ?
Modelling for the killing: YES
- ready for data from the staining assay