IGEM:IMPERIAL/2008/Prototype/Wetlab
<html> <style type="text/css"> .firstHeading {display: none;} </style> </html> <html> <style type="text/css">
table.calendar { margin:0; padding:2px; }
table.calendar td { margin:0; padding:1px; vertical-align:top; } table.month .heading td { padding:1px; background-color:#FFFFFF; text-align:center; font-size:120%; font-weight:bold; } table.month .dow td { text-align:center; font-size:110%; } table.month td.today { background-color:#3366FF } table.month td {
border:2px; margin:0; padding:0pt 1.5pt; font-size:8pt; text-align:right; background-color:#FFFFFF; }
- bodyContent table.month a { background:none; padding:0 }
.day-active { font-weight:bold; } .day-empty { color:black; } </style> </html>
| <html><a href=http://openwetware.org/wiki/IGEM:IMPERIAL/2008/Prototype><img width=50px src=http://openwetware.org/images/f/f2/Imperial_2008_Logo.png></img</a></html> | Home | The Project | B.subtilis Chassis | Wet Lab | Dry Lab | Notebook |
|---|
<html> <style type="text/css"> div.Section { font:11pt/16pt Calibri, Verdana, Arial, Geneva, sans-serif; }
/* Text (paragraphs) */ div.Section p { font:11pt/16pt Calibri, Verdana, Arial, Geneva, sans-serif; text-align:justify; margin-top:0px; margin-left:30px; margin-right:30px; }
/* Headings */ div.Section h1 { font:22pt Calibri, Verdana, Arial, Geneva, sans-serif; text-align:left; color:#3366FF; }
/* Subheadings */ div.Section h2 { font:18pt Calibri, Verdana, Arial, Geneva, sans-serif; color:#3366FF; margin-left:5px; }
/* Subsubheadings */ div.Section h3 { font:16pt Calibri, Verdana, Arial, sans-serif; font-weight:bold; color:#3366FF; margin-left:10px; }
/* Subsubsubheadings */ div.Section h4 { font:12pt Calibri, Verdana, Arial, sans-serif; color:#3366FF; margin-left:15px; }
/* Subsubsubsubheadings */ div.Section h5 { font:12pt Calibri, Verdana, Arial, sans-serif; color:#3366FF; margin-left:20px; }
/* References */ div.Section h6 { font:12pt Calibri, Verdana, Arial, sans-serif; font-weight:bold; font-style:italic; color:#3366FF; margin-left:25px; }
/* Hyperlinks */ div.Section a {
}
div.Section a:hover {
}
/* Tables */ div.Section td { font:11pt/16pt Calibri, Verdana, Arial, Geneva, sans-serif; text-align:justify; vertical-align:top; padding:2px 4px 2px 4px; }
/* Lists */ div.Section li { font:11pt/16pt Calibri, Verdana, Arial, Geneva, sans-serif; text-align:left; margin-top:0px; margin-left:30px; margin-right:0px; }
/* TOC stuff */ table.toc { margin-left:10px; }
table.toc li { font: 11pt/16pt Calibri, Verdana, Arial, Geneva, sans-serif; text-align: justify; margin-top: 0px; margin-left:2px; margin-right:2px; }
/* [edit] links */ span.editsection { color:#BBBBBB; font-size:10pt; font-weight:normal; font-style:normal; vertical-align:bottom; } span.editsection a { color:#BBBBBB; font-size:10pt; font-weight:normal; font-style:normal; vertical-align:bottom; } span.editsection a:hover { color:#3366FF; font-size:10pt; font-weight:normal; font-style:normal; vertical-align:bottom; }
- sddm {
margin: 0; padding: 0; z-index: 30 }
- sddm li {
margin: 0; padding: 0; list-style: none; float: center; font: bold 12pt Calibri, Verdana, Arial, Geneva, sans-serif; border: 0px }
- sddm li a {
display: block; margin: 0px 0px 0px 0px; padding: 0 0 12px 0; background: #33bbff; color: #FFFFFF; text-align: center; text-decoration: none; }
- sddm li a:hover {
border: 0px }
- sddm div {
position: absolute; visibility: hidden; margin: 0; padding: 0; background: #33bbff; border: 1px solid #33bbff } #sddm div a { position: relative; display: block; margin: 0; padding: 5px 10px; width: auto; white-space: nowrap; text-align: left; text-decoration: none; background: #FFFFFF; color: #2875DE; font: 11pt Calibri, Verdana, Arial, Geneva, sans-serif } #sddm div a:hover { background: #33bbff; color: #FFFFFF } </style></html>
Team Organisation
We have decided to split our wet lab work into six sub-teams:
- Cloning Strategy: Tom & Chris
- PCR: Tom & Chris
- B.subtilis Work: James
- Calibration curves
- Testing and characterisation of parts and devices
- Mircoscope co-ordinator: Krupa
Microscope information requested is to be sent to Mr. Martin Spitaler every Friday.
Cloning Strategy
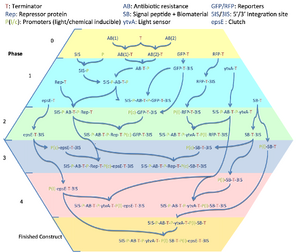
The diagram shows the critical pathway for our cloning strategy; the cloning steps we will undertake to produce our final constructs (and on the way, to produce some valuable test constructs).
We aim to re-use as many parts as possible to reduce the number of steps. However, due to planned redundancy, we often have multiple variants for some of the steps. For example, for each addition of a promoter, there are actually four cloning procedures; one for each combination of the two promoters and RBSs we have ordered. In total, we believe there are 101 cloning steps.
Without the sequences we ordered from Geneart, we can make a start by using the parts acquired from the Registry. These experiments are thus termed Phase 0. Phase 1 requires some of our Geneart sequences - most notably the promoters/RBSs - and is focused on characterisation of the constitutive promoters. Phases 2 and 3 involve construction of test constructs to characterise our inducible promoters (which will lead our epsE and biomaterial genes). Phase 4 looks at light-inducible promoters.
By the end of Phase 4, we hope to have some useful test constructs that will allow us to perform motility assays and examine the biomaterial production efficiency. If we have time (and it is feasible), an extra 4 cloning steps will allow us to produce a complete construct with all our devices placed together. If we can integrate this successfully, we will be able to perform system-level testing and maybe even produce some patterns on plates!
All the constructs have been assigned codes. The schematic for production has been finalised and can be accessed here.
Testing Constructs
Click here to view our test constructs.
Protocols
Protocol Sandbox
General Format
Link to the protocols we need!
1.Transformation Protocols for integration vectors in Bacillus subtilis
2. Transformation Protocol for linear DNA in Bacillus subtilis
3.Transformation Protocol E.coli
4. Motility Assays
5. PCR
6.Calibration Curves
7. Promoter-RBS Testing Protocols
8. Biomaterial Protocols
- Expression and Secretion
- Test on gel, looking for external product..?
- Structure formation
- Harder. Either straight binding to our product for an assay (e.g. Congo red binds to EAK16's aggregation structure) or tag our product (with e.g. a 6His tag) and assay via binding to that structure.
- Congo Red: "Our findings demonstrate that the d-EAK16 can indeed undergo self-assembly to form nanofibers and membrane at 20°C. A transparent membrane was seen when viewed under a 20X phase-contrast microscope. The membrane can be stained by Congo red dye that preferentially stains beta-sheet." [1] Widely-used method is Puchtler's Congo red stain method which can be found here (link to source paper here), although some improvements have been suggested and there exist modifications of the technique which may be better. Remember to Google "Verhoeff's Technique" when looking into this.
- His tag is 18bp long only, so shouldn't affect secretion or folding of the product when produced as a fusion protein. There are chemicals available that stain His tags (like this Invitrogen one) which would come with everything you needed and their own protocols (I'll try and find a copy online anyway).
- Harder. Either straight binding to our product for an assay (e.g. Congo red binds to EAK16's aggregation structure) or tag our product (with e.g. a 6His tag) and assay via binding to that structure.
These will collapse into the names on the official wiki...
Protocol 1
The actual protocol
Protocol 2
Second protocol
Shopping List
Reagents and Materials
Link to the Shopping list for Reagents and Materials.
Geneart
Click here to see the constructs ordered from Geneart
<html>
|
|
|