IGEM:Harvard/2006/DNA nanostructures/Notebook/2006-7-14
From OpenWetWare
Jump to navigationJump to search
Container v3.2
Pre-working stocks
- Mix 10 μL of each 50 μM stock oligo
| pre-working stock | desc | oligos plate locations | total |
| c3.2.1 | core barrel | 3.2p1A01–3.2p1H10 | 94 |
| c3.2.2 | core top lid | 3.2p2A01–3.2p2C04 | 28 |
| c3.2.3 | core bottom lid | 3.2p2C05–3.2p2E10 | 30 |
| c3.2.4 | barrel at inside apatamer locations -aptamers | 3.2p2E11–3.2p2F01 | 3 |
| c3.2.5 | barrel at outside aptamer locations -aptamers | 3.2p2F02–3.2p2F04 | 3 |
| c3.2.6 | barrel at inside apatamer locations +aptamers | 3.2p2F05–3.2p2F07 | 3 |
| c3.2.7 | barrel at outside aptamer locations +aptamers | 3.2p2F08–3.2p2F10 | 3 |
| c3.2.8 | barrel at latch locations -latches | 3.2p2F11–3.2p2F12 | 2 |
| c3.2.9 | lid at latch locations -latches (empty) | 0 | |
| c3.2.10 | barrel at latch locations +latch1 +latch2 | 3.2p2G01–3.2p2G02 | 2 |
| c3.2.11 | latch from barrel +latch1 -latch2 | 3.2p2G03–3.2p2G04 | 2 |
| c3.2.12 | latch from barrel -latch1 +latch2 | 3.2p2G05–3.2p2G06 | 2 |
| c3.2.13 | lid at latch locations +latch1 +latch2 (empty) | 0 | |
| c3.2.14 | latch from lids +latch1 -latch2 | 3.2p2G07–3.2p2G08 | 2 |
| c3.2.15 | latch from lids -latch1 +latch2 | 3.2p2G09–3.2p2G10 | 2 |
| c3.2.16 | latch staples +latch2 | 3.2p2G11–3.2p2H02 | 4 |
| c3.2.17 | displacement strands latch1 | 3.2p2H03–3.2p2H06 | 4 |
| c3.2.18 | displacement strands latch2 | 3.2p2H07–3.2p2H10 | 4 |
Make c3.2 working stocks
Final concentration of each oligo in working stocks is 250 nM.
| working stock | description | pre-working stocks (according to chart) | water | total |
| c3.2.A | -latches -aptamers |
1 (94 μL), 2 (28 μL), 3 (30 μL), 4 (3 μL), 5 (3 μL), 8 (2 μL), 9 (0 μL) | 40 μL | 200 μL |
| c3.2.B.core | +latch1 -aptamers |
1 (94 μL), 2 (28 μL), 3 (30 μL), 4 (3 μL), 5 (3 μL), 10 (2 μL), 13 (0 μL) | 40 μL | 200 μL |
| c3.2.B.latches | +latch1 -aptamers |
11 (2 μL), 14 (2 μL) | - | - |
| c3.2.C.core | +latch2 -aptamers |
1 (94 μL), 2 (28 μL), 3 (30 μL), 4 (3 μL), 5 (3 μL), 10 (2 μL), 12 (2 μL), 13 (0 μL), 15 (2 μL) | 36 μL | 200 μL |
| c3.2.C.latches | +latch2 -aptamers |
16 (4 μL) | - | - |
c3.2 Folding Experiment
Reagents
- 9 μL p7308 scaffold = (10 nM)/(44 nM) * 40 μL
- 16 μL oligos (3.2.A) = (100 nM)/(250 nM) * 40 μL
- 4 μL 10x folding buffer (500 mM HEPES pH 7.5, 500 mM NaCl, 100 mM MgCl2)
- 11 μL dH2O
- total volume: 40 μL
Annealing protocol
- start at 80[[:Category:{{{1}}}|{{{1}}}]]
- 60 cycles: wait 2 minutes, decrease 1[[:Category:{{{1}}}|{{{1}}}]]
- hold at 4[[:Category:{{{1}}}|{{{1}}}]]
Gel analysis
- 2% agarose gel supplemented to 10 mM MgCl2
- run in 1x TBE supplemented to 10 mM MgCl2
- 35 min at 130 V
- 15 min in 1x TBE, 10 mM MgCl2, 100 μg/mL EtBr (forgot to put it in the gel)
| Lane | Contents | Loading Buffer (10x TBE/glycerol) |
| 1 | 1kb DNA ladder (10 μL) | 1.1 μL |
| 2 | control: -scaffold +oligos (10 μL 25 μM) | 1.1 μL |
| 3 | control: +scaffold -oligos (10 μL 23 μM) | 1.1 μL |
| 4 | folded nanotubes (10 μL) | 1.1 μL |
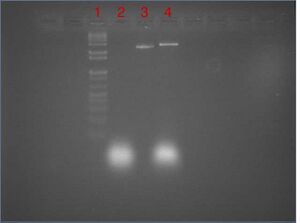
- results
- ladder, scaffold, and oligos all appear as expected
- folding reaction mixture shows oligo smear (expected) as well as a band that runs slower than the scaffold