Griffin:Lentivirus Technology
Lentiviral Transduction
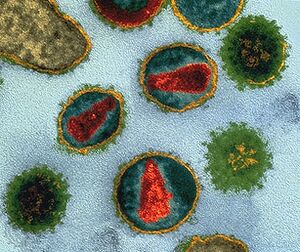
Lentivirus belonging to the Retroviridae family are ~80-100 nm diameter spherical particles with envelope/coat (YELLOW) containing an array of ~8 nm spikes (GREEN) capable of recognizing/infecting a host cell, then additional components mediate integration of viral-borne code (RED) into the unsuspecting host genome/DNA.
Lentiviral particles are highly efficient at infection and stable integration of the shRNA into a cell system. To obtain the lentiviral particle, the transfer vector that contains the shRNA cassette is already flanked by LTRs and the Psi-sequence of HIV. LTRs are necessary to integrate the shRNA cassette into the genome of the target cell, just as the LTRs in HIV integrate the dsDNA copy of the virus into its host chromosome. The Psi-sequence signal sequence is necessary for packaging shRNA into pseudovirus particles. Viral shell proteins are cotransfected into the packaging cell line (HEK 293T), so they are separate from the LTRs and Psi-sequences, and so are not packaged into virions. Third generation lentivirus particles for this reason (and others) are replication deficient. Lentiviral particles can infect both dividing and nondividing cells because their preintegration complex (virus “shell”) can get through the intact membrane of the nucleus of target cells.
- Lentiviral systems efficiently transduce both dividing and non-dividing cells
- Study long-term gene knockdown with stable expression
- Reproducibly transduce cell populations
- Inducible or constitutive gene knockdown
Lentiviral Technical Guide
Quick Reference
- Advise your tel#?
- Advise the original Purchase Order# or Sales Order #?
- Advise the catalog# / lot# on the vial?
- What species / cell type (cell name) is being tested?
- How many viral particles per cell (Multiplicity of Infection (MOI))?
- How many ug/ml Puromycin was added into culture medium, and at what time point post transduction?
- How is phenotype being measured (qPCR, WB, etc.)?
Measure transduction efficiency
- Transduction in what cell type?
- Primary cell or Continuous/immortal cell?
- Is this cell type known to have tropism for VSV-G coat protein?
- How is transduction efficiency measured for tropism to VSV-G?
- If a copGFP expressing Lentivirus was used to measure tropism, at what time point was transduction efficiency of the copGFP Control Lentiviral Particles or other reporter measured?
- How was the reporter gene measured? (FCM, IF, other)
Multiplicity of Infection (MOI)
X = How many uL of virus was used?
Y = How many cell count was transduced? NOTE: A hemocytometer is common for this step; originally designed for performing blood cell counts
MOI = X *(5000 particles/uL)/ Y
NOTE:HEK293T and other easy to transduce cells (MOI of 5-20), while neuronal cells,SHSY5Y, may require MOI of 10-50.
Puromycin Selection
- How many [ug/ml] puromycin is added post transduction?
- How was optimum puromycin concentration determined?
NOTE: Use the lowest concentration that kills 100% of non-transfected cells in 3-5 days from the start of puromycin selection (normal range; 1-10 ug/ml)
- Western blot, IF or Quantitative RT-PCR?
- Negative controls (empty virus, no virus)?
Lentivirus Transduction Protocol
Day 1:
- Plate continuous (immortal/transformed) cells (2.5x10^4) in 12 well dishes in 1mL of normal growth media (ie RPMI/10%FCS/penn&strep, DMEM/5%FBS/penn&strep).
Day 2:
- Rinse cells, and add back 0.5 mL normal growth medium.
- Add 0.5 uL of 1000x polybrene (hexadimethrine bromide) OR other Transduction Enhancement
- Try 3-4 different MOI; stock virus is 10e6 particles/200 ul = 5000 particles/ul. NOTE: Parameters below are to utilize a full 200uL unit of virus in order to optimize MOI (and avoid freeze thaw cycle of sensitive virus).
25,000 Cells
- MOI=1; (25,000 cells*1) /(5000 particles/ul) = 5 ul of virus
- MOI=3; (25,000 cells*3) /(5000 particles/ul) = 15 ul of virus
- MOI=6; (25,000 cells*6) /(5000 particles/ul) = 30 ul of virus
- MOI=10; (25,000 cells*10)/(5000 particles/ul) = 50 ul of virus
- MOI=20; (25,000 cells*20)/(5000 particles/ul) = 100 ul of virus
50,000 Cells
- MOI=1; (50,000 cells*1) /(5000 particles/ul) = 10 ul of virus
- MOI=3; (50,000 cells*3) /(5000 particles/ul) = 30 ul of virus
- MOI=6; (50,000 cells*6) /(5000 particles/ul) = 60 ul of virus
- MOI=10; (50,000 cells*10)/(5000 particles/ul) = 100 ul of virus
- MOI=20; (50,000 cells*20)/(5000 particles/ul) = 200 ul of virus
150,000 Cells
- MOI=0.5; (150,000 cells*0.5) /(5000 particles/ul) = 15 ul of virus
- MOI=1 ; (150,000 cells*1) /(5000 particles/ul) = 30 ul of virus
- MOI=3 ; (150,000 cells*3) /(5000 particles/ul) = 90 ul of virus
- MOI=5 ; (150,000 cells*5)/(5000 particles/ul) = 150 ul of virus
Day 3
- Rinse off virus and replace with normal growth cell media. 48 hours post transduction cells can undergo analysis and/or proceed with puromycin selection. There may be too few cells for qPCR or WB analysis at this step.
Day 4
- Add selection (1ug/mL Puromycin), and split cells into either 10 cm or T75 flasks containing selection.
- After 1 passage, in which non-infected cells die off in selection media (control wells that were not treated with virus), perform RNA isolation and PCR for target gene and/or immunoglobulin-based detection.
- Prepare LN2 stock once the mRNA or protein knockdown phenotype is observed. Passage durations may vary for stable gene knockdown. Monitor passage number and thaw stock accordingly to operate in the appropriate passage range for the desired knockdown phenotype.
Multiplicity of infection (MOI)
Multiplicity of infection (MOI) is a parameter for viral infectivity in a population of target cells. With wild type viruses, an infectious unit (IFU) refers to the smallest unit of virus capable of infection in a susceptible cell. Titer of viral suspension is the number of IFUs per unit volume.
Multiplicity of infection (MOI) is the ratio transducable virus per cell. MOI of 10=ten virus per cell (in the chamber /on the plate). Different cell types require different MOIs for successful transduction/knockdown of target gene. HEK293T cells are VSV-G lentiviral transduction (MOI of 5-20) while neuronal cells such as SHSY5Y often require higher MOIs of 10-50.
In most cases of transducing 42 different human cancer cell lines, MOI 3 yielded 50-90 % transduction efficiency with both vectors. In vivo studies with nude mouse s.c. tumor model (A549 lung cancer cells) revealed that lentiviruses were more efficient vehicles than adenoviruses when same amount of virus was used Molecular Therapy (2004) 9, S281.
200 uL containing 1e6 particles = 5,000 particles/uL
- HEK293= 5-10 particles/cell
- SHSY5Y= 10-50 particles/cell
When transducing a lentiviral construct into a cell line for the first time, a range of volume or MOI should be tested.
Cell Count:Confluency:Plate Diameter
Calculate MOI (MOI= viral titer/cell number)
(# cells) x (Desired MOI; typically 5-20) = Total # Particles
(# Particles) / (# cells) = MOI
NOTE
- Sensitivity of cells to lentivirus is MOI-dependent; the higher the MOI, the higher the transduction efficiency.
- Transduction efficiency is highest with the smallest cell number and largest viral volume eg: a thirty-fold increase in cell numbers resulted in a 53% decrease in efficiency (PMID 15291957)
Transduction Enhancement
Spin Infection / Spinoculation
An effective method for high efficiency viral transduction of suspension and adherent cells; spin infection can significantly enhance Lentivirus transduction.
NOTE Use of polybrene (4-8 ug/ml) during a spin infection can enhance transduction. Determination of polycation tolerance is relevant & usually in the range of 4-8 µg/ml for cultured cells.
NOTE Hard to tranduce adherent cells may benefit from spin infection. Trypsin-EDTA detachment may influence cell viability.
- Transduce cells in a multi-well plate or conical tube spun at 800 x g (< 2500 rpm) for 90 minutes at 33-37 C immediately following the addition of virus @ MOI 1-10.
- Either a swinging bucket centrifuge with 15 ml conical or an adaptor for spinning multi-well plates at 800 x g (< 2500 rpm) at 37C (or room temp) is necessary to spin infection.
Polybrene
Polycations such as Polybrene and protamine sulfate can increase gene transfer efficiency by facilitating interaction between negatively charged Viral particles and their target cells. 10 mg/ml, Store @ –20°C
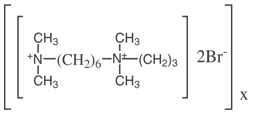
Polybrene; 1,5-dimethyl-1,5-diazaundecamethylene polymethobromide, hexadimethrine bromide.
Polybrene (hexadimethrine bromide) can increase efficiency of retroviral infection of certain cells in culture. Polybrene neutralizies the charge repulsion between virions and sialic acid on the cell surface.
Polybrene is a small positive charged molecule that binds to cell surfaces and neutralizes surface charge, increases binding between pseudoviral capsid and the cellular membrane; and greatly enhances transduction efficiency. This treatment enhances transduction of most cell types by 2-10 fold.
Concentration of Polybrene depends on cell type (usually in the range of 4-8 µg/ml).
Certain cell types (ie primary neurons, terminally differentiated neurons, and dendritic cells) are sensitive to Polybrene. Titration of polybrene using 2, 4, 6, 8 ug/ml can determine the highest nontoxic concentration. Cells can be transduced in the absence of polybrene at a higher MOI or use Protamine Sulfate.
Polybrene is highly sensitive to freeze-thaw cycles and should be stored in single-use aliquots.
Polycation Protamine Sulfate
Target cells can be infected by adding virus-containing supernatant or purified, concentrated virus diluted in media in the presence of polybrene (5ug/mL) or protamine sulfate (8ug/mL).
Cells should be infected in multi-well plates and then spun at 2500RPM for 90 minutes at 30degress immediately following addition of virus. Spin infection significantly enhances infectivity.
Fibronectin
Enhancement of the cell adhesion to the plate by pre-treating the plate with fibronectin can improve transduction.
- Fibronectin stock solution dilute in sterile PBS to 5 μg/mL
- Add 3.0 mL per well. Incubate plate(s) at 37 ºC for 1.5 hours
- Rinse each well 2× with 2.0 mL complete medium before adding cells.
- Add Cells and viral particles in complete medium containing 8 μg/mL polybrene
Puromycin Selection
Puromycin dihydrochloride is a aminonuclease antibiotic that inhibits protein synthesis. Puromycin is used for selection and maintenance of cell lines expressing a transfected pac gene (S. alboniger), whose product, puromycin acetyltransferase, inactivates puromycin via acetylation; recommended concentration in cell culture 1-10µg/ml. Lot-to-lot variations in potency exist for all selection antibiotics, each new lot of puromycin should be titrated. The working puromycin concentration for mammalian cell lines ranges from 1-10 µg/ml. Prior to using the puromycin antibiotic, titrate the selection agent to determine the optimal concentration for target cell line. Use the lowest concentration that kills 100% of non-transfected cells in 3-5 days from the start of puromycin selection.
Depending on individual cell type and doubling rate, selection of stable transfectants will take between 7 and 28 days. Expansion and characterization of single cell clones will take several weeks in addition. Media should be changed out every 2-3 days. This eliminates potentially toxic substances produced by dying cells and secondly, it keeps the concentration of the antibiotic at a constant level.
- Approximately every 2-3 days post transduction, aspirate and replace with freshly prepared selective media.
- Monitor the cells daily. Puromycin selection requires a minimum of 48 hours.
- Optimum effectiveness should be reached within 3-10 days.
Suggested working conditions for selection in some mammalian cells:
- Hela human uterus: 3 µg/ml
- HEK293 human embryonic kidney: 3
- B16 mouse melanoma: 1-3
- PC1.0 hamster adenocarcinoma: 10
- HEK293: 2
- A431: 1
- Jurkat: 4
Puromycin N-acetyl-transferase
- LOCUS STMPAC 906 bp DNA
- DEFINITION Streptomyces alboniger puromycin N-acetyltransferase (pac) gene, complete cds.
- ACCESSION M25346
- VERSION M25346.1 GI:763524
- KEYWORDS puromycin N-acetyltransferase.
- SOURCE Streptomyces alboniger
Transduction Recommendations
- Perform transductions in triplicate to minimize variability among treatment groups.
- Include positive and negative controls in each experiment.
- Seed the same number of cells in each well.
- Transduce in the presence of low level serum (0.5%) in the assay medium. Low serum can minimize signaling pathways that lead to cross-talk and high background.
- Stem cell transduction use Y-27632, also known as Rho-Associated Coil Kinase (ROCK) inhibitor, increases the cloning efficiency of human embryonic stem cells (hESCs). It prevents apoptosis and enhances the survival of single hESCs without affecting their pluripotency, or causing karyotypic abnormalities.
shRNA Transfer Vector Overview
Short hairpin RNAs (shRNAs) are typically modeled after miRNA hairpin precursors and cloned into a transfer vector that is suitable for either direct transient transfection of the shRNA, or for packaging into a lentiviral particle. Lentiviral transfer vectors are ~8-10 kb in length.
Transient shRNA Transfection
Chemically synthesized siRNA are proven and effective. The shRNA transfer vector alone can be transiently introduced into the dividing cell where the shRNA is driven by type III POL promoters, and then processed to siRNA by cellular machinery. Transient transfection is advantageous for fast analysis of shRNA mediated effects, and may be considered a preliminary approach to evaluating lentivirus dependent-transduction of shRNA expression.
Antibiotic Selection
There will be limited passage number in which "stable" antibiotic selectable lineage can be established. The gene (mRNA) must be monitored by qPCR or other method (protein) from the culture in order to determine the durability of the knockdown over passage, even if under antibiotics. shRNA targets native gene derived mRNA by recruiting host machinery in an antibiotic selectable format. Selectable antibiotic resistance is driven by (Pol II type (ie CMV) promoter), and does not control shRNA expression.
Lentivirus Dependent Stable shRNA Transduction
Viral delivery of shRNA is a powerful alternative to transfection for primary cells, and other cell types resistant to cationic lipids. Stable expression is achieved by cassette integration into the target cell chromosomes. Initially the shRNA of interest has to be introduced into the cell by viral trasnduction, subsequently into the nucleus, and then integration into chromosomal DNA.
Vector Dependent Stable shRNA Transfection
An shRNA transfer vector may express exogenous/episomal, or integrate into a genome of the target cell by antibiotic selection alone. This process may occur randomly, possibly via DNA repair/recombination. If this phenomenon does occur, integration may result in antibiotic resistance with little or no shRNA expression in passage. Integration may allow for shRNA expression. Random integration could also lead to silencing of the shRNA cassette. Several strategies may overcome negative position effects of random integration: Site-specific, homologous and transposon-mediated integration strategies are used but require the expression of integration enzymes or additional sequences on the donor template.
Nucleofection is a non-viral method of introducing DNA molecules efficiently into the nucleus of dividing cells, therefore significantly increasing the chances of chromosomal integration of the transgene. The technology was pioneered by Amaxa
shRNA Transfer Vector Promoters
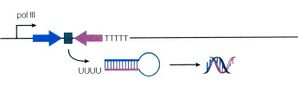
RNA Polymerase III type 3 promoters (H1, 7SK, U6) are common in vector-based expression because they are permissive expressers, they do not require any additional sequence elements, and have simple termination signals. The most abundant cellular RNAs transcribed from a type III RNA pol III promoter are the U6 small nuclear RNA (which play a crucial role in the processing of premature RNA); the 7SK RNA (a negative regulator of RNA polymerase II elongation factor TEFb); and H1 RNA (a component of RNAse P).
Unlike the majority of RNA pol III promoters, type 3 RNA Pol III promoters do not require any additional sequence elements. In addition, termination of transcription by Pol III occurs at definitive tracts of 4–5 thymidines (T4–5) (Example below), which can be inserted downstream of shRNA coding sequences in order to ensure direct termination. These 2 specific properties enable the U6, 7SK, and H1 promoters to be ideal candidates for in vivo delivery systems of siRNAs where DNA templates can undergo transcription into small RNAs with structural features resembling active siRNA/miRNA.
In summary, Pol III promoters are relatively compact, have high activity, and require a simple poly-U tract for termination.
U6
U6 small nuclear RNA (snRNA) is an essential component of the eukaryotic spliceosomes, is unique in that it is synthesized by RNA polymerase III, while all other U-snRNAs are synthesized by RNA polymerase II. U6 genes are notable for functional upstream regulatory elements which resemble RNA polymerase II regulatory sequence motifs. Tests for both potency and adverse metabolic effects upon primary cells indicate that U6 promoter may exert toxicity relative to H1 or 7SK due in part to higher activity.
U6 snRNA Class III gene initiation
1. SNAPc (SNRNA Activating Protein complex) (also termed PBP and PTF) binds to the PSE (Proximal Sequence Element) centered approximately 55 base pairs upstream of the start site of transcription. This assembly is greatly stimulated by the Pol II transcription factors Oct1 and STAF that bind to an enhancer-like DSE (Distal Sequence Element) at least 200 base pairs upstream of the start site of transcription. These factors and promoter elements are shared between Pol II and Pol III transcription of snRNA genes.
2. SNAPc acts to assemble TFIIIB at a TATA box centered 26 base pairs upstream of the start site of transcription. It is the presence of a TATA box that specifies that the snRNA gene is transcribed by Pol III rather than Pol II.
3. The TFIIIB for U6 snRNA transcription contains a smaller Brf1 paralogue, Brf2. TFIIIB is the transcription factor that assembles Pol III at the start site of transcription. Sequence conservation predicts that TFIIIB containing Brf2 also plays a role in promoter opening.
7SK
7SK is an abundant and evolutionarily conserved small nuclear RNA discovered in the 1970s. 7SK is transcribed by RNA polymerase III from one or more genes belonging to a family of interspersed repeats in the mammalian genome (Murphy et al 1984)]. The human 7SK promoter presents a strong permissivity for the nucleotide in the +1 position and recognizes a cluster of 4 or more T residues as a termination signal. The human 7SK promoter is ideal for the production of shRNAs as it can generate high amounts of shRNAs (Czauderna et al 2003, Koper-Emde et al 2004). In a series of experiments aimed to compare the strength of the human 7SK, H1 and U6 promoters, the best silencing efficiencies of various target genes was consistently obtained with a 7SK promoter (unpublished data;Invivogen).
H1
The H1 promoter drives expression of a unique gene encoding H1 RNA, the RNA component of the human RNase P. The H1 RNA gene is transcribed by RNA polymerase III into a small RNA transcript. The human H1 promoter presents the characteristics of being unusually compact. All the essential elements for transcription, that is octamer, Staf, proximal sequence element and TATA motifs, lie within 100 bp of the 5‘ flanking region. This promoter is highly permissive for the nucleotide at the +1 position which is originally an A. The termination sequence consists of a stretch of 5 thymidines. The cleavage of the transcript after the termination signal occurs after the second uridine generating a 2 nt 3‘ overhang. The H1 promoter has been successfully used in different plasmids to create siRNAs.
CMV
CMV is a RNA Polymerase II promoter. This strong promoter is active in a broad range of cell types and performs better than most pol III promoters under long term selection. However, there are specific reports that CMV driven shRNA do not function properly in certain cell types, including CD34+ hematopoietic progentior cells. Ambion offers pSilencer™ 4.1-CMV plasmid vectors designed for gene silencing experiments in a broad range of cell lines (Cat# AM5775).
The CMV promoter is considered to be a stronger promoter than other common RNA pol II promoters used in mammalian expression vectors such as Simian virus-40 (SV40) and Rous sarcoma virus (RSV) (Foecking, 1986). In vivo, RNA pol II is primarily responsible for transcription of mRNA within the cell. The CMV promoter has the advantage of being highly active in a broad range of cell types, and it is suggested to not interfere with other transcription events as may be the case with the RNA pol III U6 and H1 promoters in some situations.
shRNA Transfer Vector Components
Additional cassettes that are either essential or may be added to the lentiviral transfer vector in order to improve functionality.
AmpR
Ampicillin resistance gene for bacterial selection when growing the lentivector; ampicilin-resistance gene for selection in E.coli cells.
CMV
CMV promoter typically drives expression of copGFP (fluorescent reporter), puromycin-N-acetyl transferase (drug selectable marker) or truncated H2Kk protein (cell surface marker) for detection and selection of transduced cells.
Alternatives to CMV Promoter
Each type of promoter for selection markers offers different expression levels with different stabilities in each cell line. CMV promoter yields strong expression in many cell lines, however may not function in certain cells, including leukocytes, certain mouse cells, and stem cells. The human phosphoglycerate kinase (hPGK) eukaryotic promoter is ideal for puromycin selection when transduction of cell types known to silence CMV promoter (stem cells, leukocytes, certain mouse cells).
copGFP
CopGFP (Pontellina plumata (Arthropoda; Copepoda)) is a green fluorescent protein (excitation/ emission 482nm/502nm) as a selection marker or reporter gene.
Maximum emission wavelength: 502 nm Maximum excitation wavelength: 482 nm
- CopGFP is a novel fluorescent protein, derived from copepod plankton
- CopGFP
- CopGFP sequence
cPPT
Central polypurine tract is responsible for importing HIV provirus into the nucleus. The addition of this element between the LTRs improves integration kinetics for slow/nondividing cells. Incorporation of cPPT and a posttranscriptional regulatory element (PRE) into lentivirus vectors can improve transduction efficiency and transgene expression.
EF1
gag
Packaging signal
Biosafety Feature
None of the HIV-1 genes (gag, pol, rev) will be present in the packaged viral genome, as they are expressed from packaging plasmids lacking packaging signal--therefore, the lentiviral particles generated are replication-incompetent.
Hairpin loop
A hairpin loop sequence between sense and antisense portion. The 9-nt loop sequence (5'-TTCAAGAGA-3') is commonly used in RNA silencing experiments. A 12-nt sequence (5'-CTTCCTGTCAGA-3') may also generate similar results. Loop sequences of 3 to 15 nucleotides have been used successfully by different investigators.
Cloning Strategy for the pSIH lentiviral transfer vector
hPGK
The human phosphoglycerate kinase eukaryotic promoter is ideal for puromycin selection when transduction of cell types known to silence CMV promoter (stem cells, leukocytes, certain mouse cells).
IRES
Internal Ribosome Entry Site (IRES) sequence allows translation initiation within a messenger RNA (mRNA) sequence. IRES-containing mRNAs can undergo translation independent of regulatory mechanisms controlling recruitment of mRNA to a translation apparatus, including 5'-terminal structures; 7mG cap; methylated 5'-terminal cap structures. Efficient binding of mRNA to ribosomes is dependent on the presence of 5'-terminal 7mG. Removal of m7G decreases translation.
Pol
- Pol gene encodes for the viral protease (PRO), reverse transcriptase (RT) and integrase (IN), enzymes essential for viral replication.
Post transduction virion-bound matrix and capsid proteins disassemble, viral genome released, viral proteins reverse transcriptase (RT), integrase (IN), and Vpr proteins into the cytoplasm.
Integrase
pro-viral DNA imported into nucleus and integrated into the host genome via the action of viral integrase.
LTRs
Long terminal repeats flank either side the viral read that integrates into a host genome. 5' and 3' LTR's promote transcription (5') and polyadenylation (3') of virion RNA. 5'/3' LTR contain cis-elements necessary for viral replication (gene expression: Enhancer, promoter, transcription initiation, transcription terminator and polyadenylation signal)
- HIV-1 LTR (634 bp) consists of three regions: U3 (unique, 3′ end), R (repeated), and U5 (unique, 5′ end).
- LTRs = three regions: U3, R, and U5. U3 = enhancer and promoter sequences for viral transcription. R domain = 5′ capping sequences (5′ cap) and the polyA (pA) signal.
- Reverse transcription places two identical LTRs (U3, R, and U5 regions) at either end of the proviral DNA.
- 5'LTR = promoter for the retroviral genome, 3′LTR= viral RNA polyadenylation of the host integrated viral genome.
- 3'LTR = mediates transcription termination and polyadenylation.
Biosafety Feature
- HIV1 provirus transcription polyadenylation signals present in 3'LTR mediate in cis/viral transcript polyadenylation.
- U3 3'LTR = enhancer and promoter sequences for viral transcription.
- Deletion in the enhancer of U3 region of 3’LTR ensures self-inactivation of lentiviral construct after transduction and integration into genomic DNA of the target cells.
Psi
Retroviral Psi packaging element is a cis-acting RNA element identified in the genomes of the retroviruses Human immunodeficiency virus (HIV) and Simian immunodeficiency virus (SIV).
pUC ORI
Allows for high-copy replication in E. coli
Puro
Puromycin Resistance gene for selection of transduction events in mammalian systems.
RRE
Rev response element binds gag and involved in packaging of viral transcripts
RSV
Murine Rous sarcoma virus enhancer; Hybrid RSV promoter-R/U5 long terminal repeat; required for viral packaging and transcription. Hybrid RSV-5’LTR promoter provides a high level of expression of the full-length viral construct in 293 cells.
Biosafety Feature
The RSV promoter upstream of the 5’LTR in the lentivirus expression vector allows efficient Tat-independent production of viral RNA, reducing the number of genes from HIV-1 that are used in the packaging system. For third generation lentivirus, the trans-acting function of Tat becomes dispensable if part of the upstream LTR in the transfer vector construct is replaced by constitutively active promoter sequences.
SV40 ORI
Allows for episomal replication of plasmid in eukaryotic cells; SV40 origin for stable propagation of the pSIH plasmid in 293 producer cells.
SV40 Poly-A
Transcription termination and polyadenylation
T2A
Coexpression of a reporter gene (copGFP) and a selection marker (puro) from the same promoter (CMV) can take place with the utilization of Thosea asigna virus 2A translational cleavage site. The T2A cleavage site is ~20 amino acids long and is positioned in between the 2 transgenes (copGFP and Puro).
Cotranslational cleavage occurs via a co-translational ribosome skipping mechanism between the C-terminal Glycine and Proline residues, leaving 17 residues attached to the end of copGresidue to the start of puromycin resistance gene.
This allows for balanced expression/coexpression of a reporter gene and a resistance gene/cDNA from the same promoter (CMV).
WPRE
Woodchuck hepatitis virus post-transcription regulatory element is capable of
- Prevents poly A site readthrough
- Promotes RNA processing and maturation
- Increases nuclear export of RNA
- For genomic transcripts, enhances vector packaging and increases titer.
- For transduced cells, WPRE stabilizes and improves nuclear export of shRNA transcripts.
qPCR lentiviral titration with WPRE
shRNA Transfer Vector Abbreviated Position pSIH-H1
- RSV/5'LTR 7-414 Hybrid RSV promoter-R/U5 long terminal repeat; required for viral packaging and transcription.
- gag 567-919 Packaging signal.
- RRE 1076-1309 Rev response element binds gag and involved in packaging of viral transcripts.
- cPPT 1798-1916 Central polypurine tract (includes DNA Flap region) involved in nuclear translocation and integration of transduced viral genome.
- CMV promoter 1922-2271 Human cytomegalovirus (CMV)--constitutive promoter for transcription of puromycin.
- Puro 2279-2878 Puromycin-resistant marker for selection of the transfected/transduced cells.
- WPRE 2885-3425 Woodchuck hepatitis virus posttranscriptional regulatory element--enhances the stability of the viral transcripts.
- 3' ∆LTR (∆U3) 3564-4038 Required for viral reverse transcription; self-inactivating 3' LTR with deletion in U3 region prevents formation of replication-competent viral particles after integration into genomic DNA.
- H1 RNA promoter 3602-3818 RNA polymerase III promoter for expression of siRNA insert.
- SV40 Poly-A 4110-4241 Transcription termination and polyadenylation.
- SV40 Ori 4250-4396 Allows for episomal replication of plasmid in eukaryotic cells.
- pUC Ori 4766-5439 (C) Allows for high-copy replication in E. coli.
- AmpR 5584-6444 (C) Ampicillin resistant gene for selection of the plasmid in E. coli.
Lentiviral Particle Assembly
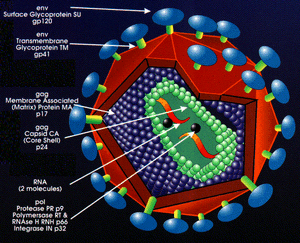
Lentiviral (VSV-G) pseudotype particles are created in a transient transfection system in which a cell line (HEK293T) is co-transfected with 3-4 separate plasmids. These include a transfer vector plasmid (cPPT & shRNA cassette), a packaging plasmid, and a plasmid with the heterologous envelop gene (ENV) of a different virus (VSV-G). Pseudoviral particles are collected from culture media and must undergo titration to measure infectivity prior to experimental use.
Reverse transcriptase and viral integrase components are packaged within the viral particles for cytosolic RT, nuclear translocation, and integration.
SCBT Inc. Lentivirus technology utilize third generation technology for biosafety level 2 (BSL2) compliant (plasmid) expression. Lentiviral VSV-G pseudotype particles can be produced from shRNA transfer vectors within third generation viral packaging kits, in which a packaging cell line (HEK293T) is co-transfected with 3-4 separate plasmids as part of a pseudotype viral packaging system. SCBT Inc. shRNA transfer vectors are suitable for viral packaging.
Transfer Vector Plasmid
The transfer vector plasmid (~8-10 kb) contains cis-acting genetic sequences necessary for the vector to infect the target cell and for transfer of the shRNA cassette, and contains restriction sites for insertion of the shRNA. The 3’ and 5’ LTRs, the original envelop proteins, and gag sequence promoter have been removed. The shRNA transfer vector alone can be transiently introduced into the dividing cell where the shRNA is synthesized by cellular machinery.
In order to culture the transfer vector effectively and avoid recombination events, a suitable e. coli strain Invitrogen MAX Efficiency® Stbl2™ Competent Cells SKU# 10268-019, and culture temperature; 30°C expression, incubation temperature is important to optimize performance of the competent cells to produce intact lentivector.
However, there are distinct differences between E. Coli-dependent vector-based recombination events versus point mutations. Both can influence the fidelity of the lentivector, however the implications of point mutations in the construct is much more significant. Since the labor costs associated with cloning and production of the shRNA constructs are significant, be sure to invest your time in a transfer vector that is proven in literature or contains lot specific data for adequate controls from the vendor. DO NOT assume the vector you are purchasing on paper is the one you receive in the vial.
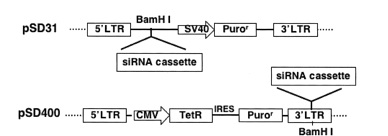
lentivector pSD31 for constitutive shRNA expression and pSD400 for conditional shRNA expression
Constitutive transfer vectors
Currently, most siRNA expression vectors are engineered to drive siRNA transcription from the polymerase III promoters U6 and H1. These promoters are particularly suited for hairpin siRNA (shRNA) expression, since they contain all of the cis-acting promoter elements upstream of the transcription initiation site and deploy a polyT transcription termination site that leads to the addition of 2-nucleotide (nt) overhangs (UU) to shRNAs, a feature that is important for siRNA function.
- Zhang J, Wang C, Ke N, Bliesath J, Chionis J, He QS, Li QX, Chatterton JE, Wong-Staal F, and Zhou D. A more efficient RNAi inducible system for tight regulation of gene expression in mammalian cells and xenograft animals. RNA. 2007 Aug;13(8):1375-83. DOI:10.1261/rna.520707 |
- Paule MR and White RJ. Survey and summary: transcription by RNA polymerases I and III. Nucleic Acids Res. 2000 Mar 15;28(6):1283-98. DOI:10.1093/nar/28.6.1283 |
Inducible (conditional) transfer vectors
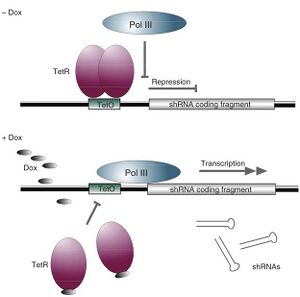
Tetracycline Operator
Specific inhibition of gene expression using a stably integrated siRNA (Tiscornia et al. 2003) is not suitable for silencing genes that are involved in cell growth or survival (Zhou et al. 2006). In such cases, regulating siRNA expression in mammalian cells has become essential. Having the ability to control when and how much of a particular siRNA is expressed makes it possible to study temporal- and concentration-dependent effects on a host system and allow cells to grow prior to gene silencing by “toxic” siRNAs. Furthermore, studying a host cell's response to the presence and absence of siRNA often leads to further understanding of the target gene's function.
The inducible RNAi system uses a modified form of the regulated, tetracycline-controlled gene expression system described by Gossen and Bujard (2002) in mammalian cells for the purpose of temporal silencing target genes. This system relies on two components: a tetracycline regulatory protein (TetR), which has affinity to the tetracycline operator (TetO); and a TetO-tethered pol III promoter, whose transcription activity is blocked by binding to the regulatory TetR. In the absence of tetracycline (Tet), the TetR protein binds to the TetO sequence within the promoter and acts as a potent transcription suppressor. The shRNA expression is restored when the cell culture medium is added with either Tet or doxycycline (Dox), a Tet derivative, which binds to and causes dissociation of TetR from the pol III promoter via a TetR conformation change (Zhou et al. 2006). This system allows for observation of the loss-of-function phenotypes under noninduced and induced conditions in the same isogenic cells, excluding other potential interferences.
- Zhang J, Wang C, Ke N, Bliesath J, Chionis J, He QS, Li QX, Chatterton JE, Wong-Staal F, and Zhou D. A more efficient RNAi inducible system for tight regulation of gene expression in mammalian cells and xenograft animals. RNA. 2007 Aug;13(8):1375-83. DOI:10.1261/rna.520707 |
- Qin XF, An DS, Chen IS, and Baltimore D. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc Natl Acad Sci U S A. 2003 Jan 7;100(1):183-8. DOI:10.1073/pnas.232688199 |
- Tiscornia G, Singer O, Ikawa M, and Verma IM. A general method for gene knockdown in mice by using lentiviral vectors expressing small interfering RNA. Proc Natl Acad Sci U S A. 2003 Feb 18;100(4):1844-8. DOI:10.1073/pnas.0437912100 |
- Gossen M and Bujard H. Studying gene function in eukaryotes by conditional gene inactivation. Annu Rev Genet. 2002;36:153-73. DOI:10.1146/annurev.genet.36.041002.120114 |
- Zhou D, He QS, Wang C, Zhang J, and Wong-Staal F. RNA interference and potential applications. Curr Top Med Chem. 2006;6(9):901-11. DOI:10.2174/156802606777303630 |
- Herold MJ, van den Brandt J, Seibler J, and Reichardt HM. Inducible and reversible gene silencing by stable integration of an shRNA-encoding lentivirus in transgenic rats. Proc Natl Acad Sci U S A. 2008 Nov 25;105(47):18507-12. DOI:10.1073/pnas.0806213105 |
Envelope Gene Plasmid
The envelope coat protein (pseudotype) of a virus specifies tropism.
Envelope (viral coat) proteins
Vesicular stomatitis virus G glycoprotein (VSV-G)
VSV is broadly tropic and highly efficient at particle production. Broad tropism. Less inflammatory with in vivo tropism for the liver. Pseudotyping viral vectors with vesicular stomatitis virus glycoprotein (VSV-G) enables the transduction of an extensive range of cell types from different species. Two important parameters of the VSV-G-pseudotyping phenomenon relating to the transduction potential of lentiviral vectors: (1) the glycosylation status of VSV-G, and (2) the quantity of glycoprotein associated with virions.
- Burns JC, Friedmann T, Driever W, Burrascano M, and Yee JK. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc Natl Acad Sci U S A. 1993 Sep 1;90(17):8033-7. DOI:10.1073/pnas.90.17.8033 |
- Borok Z, Harboe-Schmidt JE, Brody SL, You Y, Zhou B, Li X, Cannon PM, Kim KJ, Crandall ED, and Kasahara N. Vesicular stomatitis virus G-pseudotyped lentivirus vectors mediate efficient apical transduction of polarized quiescent primary alveolar epithelial cells. J Virol. 2001 Dec;75(23):11747-54. DOI:10.1128/JVI.75.23.11747-11754.2001 |
- Reiser J. Production and concentration of pseudotyped HIV-1-based gene transfer vectors. Gene Ther. 2000 Jun;7(11):910-3. DOI:10.1038/sj.gt.3301188 |
- Desmaris N, Bosch A, Salaün C, Petit C, Prévost MC, Tordo N, Perrin P, Schwartz O, de Rocquigny H, and Heard JM. Production and neurotropism of lentivirus vectors pseudotyped with lyssavirus envelope glycoproteins. Mol Ther. 2001 Aug;4(2):149-56. DOI:10.1006/mthe.2001.0431 |
- Farley DC, Iqball S, Smith JC, Miskin JE, Kingsman SM, and Mitrophanous KA. Factors that influence VSV-G pseudotyping and transduction efficiency of lentiviral vectors-in vitro and in vivo implications. J Gene Med. 2007 May;9(5):345-56. DOI:10.1002/jgm.1022 |
Packaging Plasmid
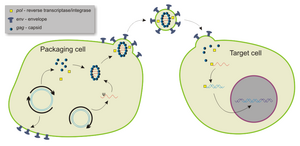
The packaging plasmid is the backbone of the virus system. In this plasmid are found the elements required for vector packaging such as structural proteins, HIV genes (except the gene env which codes for infection of T cells, or the vector would only be able to infect these cells), and the enzymes that generate vector particles.
gag/rev/pol
Most HIV lentivector systems require only 3 HIV encoded proteins, Gag, Pol, and Rev.
The RNA genome of HIV consists of at least 7 structural landmarks (LTR, TAR, RRE, PE, SLIP, CRS, INS) and nine genes (gag, pol, and env, tat, rev, nef, vif, vpr, vpu, and tev) encoding 19 proteins. gag & pol contain information needed to make the structural proteins for new virus particles. The rev protein (p19) is involved in shuttling RNAs from the nucleus and the cytoplasm by binding to the RRE RNA element.
CMV
Also contained is the human cytomegalovirus (hCMV) which is responsible for the expression of the virus proteins (gag, rev, pol) during translation. The packaging signals and their adjacent signals are removed so the parts responsible for packaging the viral DNA have been separated from the parts that activate them. Thus, the packaging sequences will not be incorporated into the viral genome and the virus will not reproduce after it has infected the host cell.
- Amado RG and Chen IS. Lentiviral vectors--the promise of gene therapy within reach?. Science. 1999 Jul 30;285(5428):674-6. DOI:10.1126/science.285.5428.674 |
- Naldini L, Blömer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, and Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996 Apr 12;272(5259):263-7. DOI:10.1126/science.272.5259.263 |
Replication Competent Virus (RCV) Testing
- SCBT Inc. Lentivirus technology utilize third generation biosafety level 2 (BSL2) compliant (plasmid) expression systems.
Most viral vectors are disabled so replication competent viruses are not readily formed by any known biological process in normal hosts. Lentivirus RCV testing is performed by ELISA for the presence of p24 antigen in the media of cell culture inoculated with viral stock solution. Confirmation of the complete absence of RCV must be documented prior to use in animals.
RCV testing is not required for third-generation, commercially available Lentiviral vector systems.
Third generation HIV-based Lentivirus expression systems were originally for gene therapy applications. Due to their biosafety (BSL2), commercial vendors including Santa Cruz Biotechnology, Inc. now offer these safe, and easy-to-use products.
3rd generation pseudoviral particles can infect target cells and express effector or reporter molecules but cannot replicate within target cells for two reasons:
1. The viral structural genes are absent
2. The LTRs are designed to be self-inactivating upon transduction
- PMID=9765382
- Santa Cruz Biotechnology Inc.
- System Biosciences
Biosafety of 3rd generation Lentivirus
The third generation of lentivirus vectors provides multiple safeguards against the production of replication competent lentivirus (RCL) (Dull, 1998) and are suitable for use under Biosafety Level 2 environment.
3rd generation Lentivirus Biosafety Features
- A deletion in the enhancer of the U3 region of 3’ΔLTR ensures self-inactivation of the lentiviral construct after transduction and integration into genomic DNA of the target cells. Long terminal repeats encompass the cassettes that incorporate into the host genome. The 5' and 3' LTR's serve to promote transcription and polyadenylation of the virion RNA's. The LTR contains all other cis-acting sequences necessary for viral replication. Deletion in the enhancer of U3 region of 3’LTR ensures self-inactivation of lentiviral construct after transduction and integration into genomic DNA of the target cells.
- The RSV promoter (in HIV-based vectors) upstream of 5’LTR in the shRNA transfer vector allow efficient Tat-independent production of viral RNA, reducing the number of genes from HIV-1 that are used in this system. For third generation lentivirus, the trans-acting function of Tat becomes dispensable if part of the upstream LTR in the transfer vector construct is replaced by constitutively active promoter sequences.
- The number of lentiviral genes necessary for packaging, replication and transduction is reduced to three (gag, pol, rev).
- The corresponding proteins are expressed from different plasmids (for HIV-based packaging plasmids) that lack packaging signals. The packaging plasmids share no significant homology to any of the expression lentivectors, the pVSV-G expression vector, or any other vector, to prevent generation of recombinant replication competent virus.
- None of the HIV-1 genes (gag, pol, rev) are present in the packaged viral genome, as they are expressed from separate plasmids lacking packaging signal. Therefore, the lentiviral particles generated are replication-incompetent. The genetic elements are split into four plasmids.
- Produced pseudoviral particles will carry only a copy of the clone-in construct.
- Lentiviral particles produced in this system are pseudotyped with envelope G glycoprotein from Vesicular Stomatitis Virus.
Production of RCV can only be the result of four unlikely events: recombination of four plasmids and reconstitution of the U3 LTR promoter activity. Since the probability of the generation of an RCV during vector production is excessively low, vector batches would be contaminated with low number of RCV particles if any (PMID:12907156). Rodents are dead end hosts for such RCV: viruses could enter into cells but not produce any progeny in vivo. Infectious virus production by cells from HIV-transgenic mice was documented only ex vivo under special conditions (PMID:14585206). RCV amplification in rodents is therefore highly unlikely.
A system incorporating all these safeguards can be seen as safe and is usually classified as BSL2.
- PMID=9765382
- Santa Cruz Biotechnology Inc.
- System Biosciences
Biosafety Level 2 (BSL-2)
BSL-2 Procedure
- Check with Institutional health and safety guidelines regarding use of lentivirus and follow standard microbiological practices.
- Wear gloves and lab coat all the time when conducting the procedure;
- All procedures are performed carefully to minimize the creation of splashes or aerosols.
- Work surfaces are decontaminated at least once a day and after any spill of viable material.
- All cultures, stocks, and other regulated wastes are decontaminated before disposal by an approved decontamination method such as autoclaving. Materials to be
decontaminated outside of the immediate laboratory are to be placed in a durable, leakproof container and closed for transport from the laboratory.
BSL-2 Spill
- 1) Alert others in the lab to evacuate due to a biological spill, then close lab entrance door
- 2) If the spilled material may be aerosolized:
- a. Post a warning sign on the entrance door (“Biohazard Spill – Do Not Enter”)
- b. Allow aerosols to settle for at least 30 minutes before starting cleanup
- c. Notify EHS if HEPA filtered respirator is required to prevent inhalation exposure
- 3) Put on appropriate personal protective equipment (gloves, lab coat, face protection, etc.)
- 4) Cover spill with absorbent material (i.e. paper towels)
- 5) Carefully pour disinfectant around edges of spill and then work inward (avoid splashing)
- 6) Allow 20 minute disinfectant contact time
- 7) Remove broken glass or other sharps with a brush and dustpan, tongs, or forceps
- a. Place contaminated sharps in a puncture-resistant biohazard sharps container
- 8) Use paper towels to wipe up spilled material, then dispose of towels with infectious waste
- 9) Wipe down all surfaces or items once more with absorbent material and disinfectant
- 10) Place all contaminated disposable materials not containing sharps in a biohazard bag
- 11) Place all contaminated re-usable items in biohazard bag, then sterilize by autoclaving
- 12) Remove gloves and other protective equipment, then wash hands with soap and water
BSL-2 Personnel contamination
- 1. Remove contaminated clothing (without exposing more skin), then place contaminated clothing in a biohazard bag for autoclaving
- 2. Thoroughly wash the exposed area of the body with soap and water
- 3. Report the incident to your supervisor or Principal Investigator
- 4. Report the incident to the Biosafety Officer for further investigation
- 5. Seek medical attention if the contamination has resulted in a potential exposure