Griffin:CRISPR
CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) Cas Nuclease
Prokaryotic CRISPR array-Cas Nuclease (CRISPR/Cas) is an adaptive RNA-guided anti-[phage, virus, plasmid, transposon, integrative/conjugative elements, genomic island] defense system. CRISPR/Cas systems are capable of supporting mammalian gene editing (genetic engineering) applications by virtue of rational CRISPR RNA (crRNA/∼20 bp) sequence design coupled with exogenous genetically modifiable Cas Nuclease expression.
Genome-scale CRISPR Knock-Out (GeCKO) CRISPR Libraries
- GeCKO sgRNA libraries target early 5' exons filtered minimize off-target modification, for genome editing.
- GeCKO v2 Knockout screening
- GeCKO v2 human library target 5′ exons of 19,050 human genes with 6 sgRNAs (3xA / 3xB) per gene. Total sgRNA 123,411 (65,383 in Library A, 58,028 in Library B)
- GeCKO v2 human library target 5′ exons of 20,611 mouse genes with 6 sgRNAs (3xA / 3xB) per gene. Total sgRNA 130,209 (67,405 in Library A, 62,804 in Library B)
- Vector capable of producing higher-titer virus (lentiCRISPRv2)
CRISPR/Cas Lentiviral (Library Screen)
- Define the library constraint (ie whole genome (whole coverage), kinases only, gene family specific, etc) with 4-8 sgRNA per gene targeted (multiplex the knockout observation).
- Determine a suitable lineage (immortal/continuous (ie multiplex similar differentiation states (ie early stage cancer stem cell))) with favorable VSV-G tropism (easy to infect), multiple passage compliant (14-30 day screening)
- Package Cripsr RNA (plasmid) library (>75000 sgRNA) into VSV-G lentivirus ---> (sgRNA library virus)
- Transduce Cas9 into target cell (immortal/continuous)
- Transduce (sgRNA library virus) @ low MOI <1 (0.3-0.5 MOI) = achieve stochiometry of <1 sgRNA sequence type per target cell.
- Select with antibiotic (puromycin) to propagate tranduction+ population to 100e6 cells (100x 10cm plates (1e6 cells/10cm))
Screen
- Day 0 (100e6 cells) screen control condition + treatment (experimental) arm for DayXX (ie 3-10 days of a therapeutic)
- Extract DNA for determination of guideRNA sequences.
- Send Day0 and DayXX to (Next Generation Sequencing NGS) to determine enriched counts for each sgRNA in the total cell populations.
- Count #sgRNA in Day0 and compare to Count #sgRNA in DayXX. Compare enriched vs depleted sgRNA.
Cancer stem cell example
- Minimum 5 sgRNA per gene to determine across the board replication of cell depletion or enrichment.
- DEPLETED Guides for driver genes will be depleted b/c cells with pro-growth genes knocked out will die.
- ENRICHED Guides for suppressor genes will be enriched b/c cells with suppressor genes knocked out will accelerate.
- R Algorithms assign a score and p value (EdgeR, DESeq2, BAGEL, MAGeCK) to each gene in order to determine cancer fitness genes and align with what pathways (Cell Cyle, Mitchondria, ER/Golgi, Cholesterol Biosynth).
- Perform mechanistic gene studies.
CRISPR Editing Evaluation
- PCR Strategies for Cas9/HDR Homology Directed Repair HDR genomic integration variability
- Tracking of Indels by DEcomposition TIDE
- TIDE (NHEJ Indel)/ TIDER (templated (HDR))
Nascent Genome Repair Mechanisms
- 3 part repair mechanism = (1st) DSB recognition (2nd) processing of nonligatable DNA termini (3rd) joining of two suitable DSBs
- NHEJ constrains broken DNA ends in the absence of end resection (MMEJ) for repair initiation (ie Insertion/Deletion (Indel)).
- NHEJ : Ku heterodimeric repair scaffolds associate with DSB ends.
Insertion/Deletion (Indel)
- Cas9/KO dependent (InDel) are useful for gene expression disruption, and observable within 12 hours at high efficiencies (<70%). Inertion Deletion (Indel) of bases in the genome of a cell can yield frame shift or premature stop codons and/or disruption of transcription.
- Cas9/KO dependent InDel introduction occurs when complimentary DNA (double strand breaks (DSB)) strands undergo micro-alignment dependent end resection (MMEJ) or repair misalignment (NHEJ) within a host genome, yielding frame shift, premature stop codon(s) (premature termination), and/or disruption of transcription.
- InDel being introduced at DSB are thought to be random
- NHEJ (non-homologous end joining) [DNA-PKcs, 53BP1, Xlf, Xrcc4 and Lig4]-dependent DNA DSB repair yields either perfect repair or (small) <10 bp insertion/deletion of bases in the host genome. NHEJ repair of Cas9-dependent double-stranded breaks generates small < 10 bp insertions and deletions (indels).
- MMEJ (microhomology-mediate end joining repair) (resection-mediated) Mre11-Rad50-Nbs1 (MRN) complex, promoted by Ctip and Brca1, (circumvents NHEJ and) resects (to cut out) a span (>16 bp) of broken/damaged/DSB DNA, (Parp1, Polϑ and ligases Lig1 and Lig3)-dependent repair.
HDR Plasmid can undergo cotransfection with Cas9/KO plasmid(s), and facilitates hereditary antibiotic-dependent cytotoxic selection of cells with genomic integration at Cas9-induced DNA cleavage sites. Homology Directed Recomination (HDR)/integration of an RFP/Puromycin resistance cassette.
SCBT Inc. HDR integration cassette includes LoxP motifs, the eukaryotic translation elongation factor 1 α (EF‐1α/EEF1A1) promoter, and both RFP and puromycin N-acetyltransferase all integrate into nascent genome at sites of RNA guided Cas9 endonuclease-dependent DSBs where 5' and 3' arm complimentarity are present.
- Overview
- Rad51 family association with DSBs recruit accessory factors directing genomic recombination via homologous arms on an exogenous repair template.
- PCR Strategies for Cas9/HDR Homology Directed Repair HDR genomic integration variability
- L-755,507 (CAS 159182-43-1) is a selective β3-adrenergic receptor partial agonist
- L755507 and Brefeldin A, achieve maximal effects at 5 μM and 0.1 μM respective; increases efficiency of HDR dependent GFP insertion ~3 fold vs. DMSO-treated. L755507 increases HDR efficiency in cell lines (K562 and HeLa), suspension cells (K562), primary neonatal cells (HUVEC and fibroblast CRL-2097), and human ES cell-derived cells (neural stem cells)
- (Li et al. 2011) Mechanistic by which L755507 enhances CRISPR-mediated HDR.
- Treating different types of cells with L755507 shows improved HDR efficiency, ~2 fold in human umbilical vein endothelial cells (HUVEC). PMID: 25658371
Cas9
Cas9 is an RNA guided DNA double strand endonuclease, that generates blunt end double strand DNA cuts via sgRNA targeting genomic DNA.
Type II CRISPR / RNA guided DNA Endonuclease / blunt end double strand DNA cut
- Blunt ends; protospacer-adjacent motif (PAM) immediately downstream (3') of the target site.
- saCas9 : 1053 aa Staphylococcus Aureus Cas9. PAM = NNGRRT. blunt end dsDNA cut
- spCas9 : 1368 aa Streptococcus Pyogenes Cas9. PAM = N20-NGG. Versatile balance between PAM complexity (specificity) and construct size.
- stCas9 : 1121 aa Streptococcus thermophilus (St1Cas9) PAM = NNAGAAW. 1388 aa St3Cas9 PAM = NGGNG
sgRNA (single guide RNA)
- ~64-nt single guide RNA (hairpin sgRNA = crRNA+tracRNA) to encode target specificity (~20nt footprint of genomic DNA complimentarity + PAM).
- Chimeric crRNA-tracrRNA hybrids (sgRNA) direct Cas9 cleavage within mammalian genomes to stimulate NHEJ or HDR-mediated genome editing.
- The first 20 nucleotides (nt) at the 5’ end are complementary to the target DNA. The remaining nt form a hairpin structure.
Cas9 Nickase
Wild-type Cas9 Nuclease cleaves DNA via RuvC and HNH nuclease domains, that generate blunt end double strand breaks. Double nicking is achievable when Cas9 nickase (RuvC / HNH) complexing with appropriately spaced target sites mimic a double strand break via cooperative nicks.
There are 2 plasmids harboring different nickases that each cut opposing strands of the genomic DNA guided by unique sgRNA respectively. and these 2 plasmids harboring GFP on one and Puromycin resistance on the other. Both transient
- Nickase HNH+ : HNH single nuclease domain cleaves DNA strand complementary to the RNA guide. Type 2 Streptococcus Cas9 contains 2 nuclease domains; RuvC / HNH
- Nickase RuvC+ : RuvC 3 subdomain (RuvC1 @ N-terminal, RuvC2/3 flank HNH). Nuclease cleaves DNA strand non-complementary to the guide RNA. Type 2 Streptococcus Cas9 contains 2 nuclease domains; RuvC / HNH
dCas9
To enable eukaryotic genome regulation, previously established transcriptional activators have been fused to a dCas9 ortholog from the bacteria Streptococcus pyogenes (dSpCas9). Catalytically inactive Cas9; deactivated (dCas9), is nuclease deficient.
- dCas9-SAM: NLS-dCas9-VP64, and MS2-p65-HSF1 trans-acting native gene synergistic activation mediator (SAM).
- dCas9-SSAP: Single-strand annealing protein(SSAP) dependent cleavage-free kilobase-scale (long-sequence genome engineering) gene editor (insertion).
Components
Cas9/KO
- U6 promoter:human
- T2A peptide:Thosea asigna virus
- GFP:Aequorea victoria
- NLS:SV40 virus
- Cas9:Streptococcus pyogenes bacteria
- CBh promoter:chicken
- gRNA scaffold:bacteria
- 20 nt non-coding RNA sequence
HDR
- EF1a promoter—> human / versus CMV promoter, eukaryotic translation elongation factor 1 α (EF‐1α/EEF1A1) promoter increases transfection efficiency, transgene expression, and proportion of expression‐positive clones/copy number. constitutively active in a broad range of cell types
- RFP:sea anemone Entacmaea
- Puromycin:bacteria Streptomyces alboniger
- T2A peptide:Thosea asigna virus
Cre Recombinase
- Promoter:CMV virus
- Cre:bacteriophage
- NLS:SV40 virus
Type V CRISPR / RNA guided DNA Endonuclease / generates staggered cuts with 5'overhang
- Cas12a: higher specificity/ lower seed mismatch tolerance (staggered dsDNA cut) versus Cas9 (blunt end dsDNA cut).
- Acidaminococcus sp. BV3L6 Cas12a (AsCas12a, AsCpf1). 5′-TTTN-3′ PAM / staggered cut @ [~19bp downstream PAM (sense)/~23bp downstream PAM (antisense)].
- Lachnospiraceae Cas12a (LbCas12a, LbCpf1).
- Francisella novicida Cas12a (FnoCas12a).
- Edits AT-rich genomes/regions/ SNP-specific editing.
- Only requires crRNA (tracrRNA-independent)/processes crRNA without tracrRNA.
- Dna Endonuclease-TargEted Crispr TRansreporter (DETECTR) achieves attomole DNA detection merging isothermal amplification with Cas12a.
- DETECTR
- 1) RNA extracted from a sample
- 2) reverse transcription (2&3 are in 1 reaction)
- 3) isothermal amplification using loop-mediated amplification (RT–LAMP)
- 4) pathogen sequence targeting gRNA dependent Cas12 detection
- 5) Activated Cas12 dependent cleavage of a reporter molecule confirms positive detection
Type VI CRISPR / RNA guided RNA Endonuclease / permissive 'collateral' RNA cleavage
- Programmable RNase activity= spacer complementarity-independent RNA/transcript degradation.
- ~64-nt guide RNA scaffold for RNA endonuclease activation/ target specificity.
- Target specificity via 28-30-nt spacer complementarity to target RNA.
- Collateral cleavage
- Collaborative post-recognition (non-specific) degradation of surrounding transcripts is limited to bacteria / not present in plant or mammal
- crRNA spacer ~24 nt stem-loop structure with a mononucleotide protospacer
- Specific High-sensitivity Enzymatic Reporter unLOCKing SHERLOCK Microbiology Diagnostic
- SHERLOCK
Cas13a (C2c2)
Cas13b (C2c6)
Cas13d
- Metabolizes endogenous transcript splicing
- Possibile in vivo delivery due small size (~ 930 aa) /smallest size for class 2 CRISPR.
dCas13
Nomenclature
- CBh : chicken β-actin (CBA) hybrid promoter. Replace the SV40 intron in the CBA promoter with a hybrid intron composed of a 5′ donor splice site from the chicken β-actin 5′ UTR and a 3′ acceptor splice site from from minute virus of mice (MVM). This new version of the chicken β-actin “promoter” (enhancer, promoter, and 5′ UTR) = “CBh” for CBA hybrid intron. 800-bp CBh promoter elicits durable expression.
- CMV : human cytomegalovirus (CMV) promoter regulatory drives constitutive protein expression; prone to methylation/silencing (ie iPSC)
- DSB : DNA double strand breaks undergo repair via error prone non-homologous end joining (NHEJ) or precision homology directed repair (HDR).
- EF1A : EF-1α promoter eukaryotic translation elongation factor 1 α (EF-1α, gene symbol EEF1A1) promoter increases transfection efficiency, the transgene expression, the proportion of expression-positive clones and in vitro copy number. Core EF1a promoter ~200-300bp in size. The ~1172bp intron-containing version (occasionally spliced out); the extra sequence yields higher level of expression than the core EF1a promoter alone.
- HNH : Cas9 Nuclease lobe (1 of 2); single nuclease domain.
- HDR : Rad51 family association with DSBs recruit accessory factors directing genomic recombination via homologous arms on an exogenous repair template.
- Indel : Insertion deletion of bases in the genome of a cell.
- NHEJ : Ku heterodimeric repair scaffolds associate with DSB ends. Stochastic insertions and deletions (Indel) introduction occurs when complimentary strands undergo micro-alignment dependent end resection / repair misalignment leading to frame shift. Useful for gene disrutpion. Observable within 12 hours at high efficiencies (<70%).
- NUC : Nuclease (NUC) lobe consisting of HNH domain, assembled RuvC subdomains, and a PAM-interacting (PI) C-terminal region.
- PAM : Target DNA sequence (protospacer) must be gRNA complimentary and contain a "protospacer adjacent motif" DNA sequence. PAM recognition -> rate-limiting R-loop propagation -> DNA dsDNA cleavage. protospacer adjacent motif (PAM) = ~2–6-base pair DNA sequence 3' to target DNA sequence of the RNA guided Cas9 nuclease (CRISPR bacterial adaptive immune system).
- REC : α-helical recognition (REC) lobe
- RGN : RNA guided Nuclease
- RuvC : Cas9 Nuclease lobe (2 of 2) RuvC contains three subdomains; RuvC I near the N-terminal region of Cas9 and RuvC II/III flanking the HNH domain near the middle of the protein.
- sgRNA : Fusion of ~20nt CRISPR RNA (crRNA) + ~60nt trans-activating RNA (tracRNA) simplifies RNA:Cas9 complex ratio from 2:1 -> 1:1, and improves editing rates relative to a dual RNA strategy. Targeted deletion utilizing 2+ sgRNA result in repair naive junctions by either sgRNA alone =higher % of Indel events. 1 sgRNA blunt end DSBs repair correct at a higher fidelity than 2+ sgRNA DSBs repair.
- Seed Region: A subset of nucleotides within the crRNA (sgRNA) that pairs with PAM-proximal nucleotides; sensitive to mismatches, necessary for target affinity and specificity. Rate of genomic target binding (not affinity) decreases with each mismatch; CRISPRcas12 crRNA contains a larger seed region (lower mismatch tolerance) versus CRISPRcas9 (∼10 bp of the RNA-DNA helix (R-loop) proximal to the PAM).
- Termination Signal: Poly TTTTTT will generate a "poly A tail" on nascent mRNA. Polyadenylation (poly(A)) signals (PAS) are a defining feature of eukaryotic protein-coding genes. The poly(A) tail is important for the nuclear export, translation and stability of mRNA.
- U6: U6 is a type III RNA polymerase III promoter commonly used for driving ncRNA expression.
GCTAGCGAGGGCCTATTTCCCATGATTCCTTCATATTTGCATATACGATACAAGGCTGTTAGAGAGATAATTGGAATTAATTTGACTGTAAACACAAAGATATTAGTACAAAATACGTGACG TAGAAAGTAATAATTTCTTGGGTAGTTTGCAGTTTTAAAATTATGTTTTAAAATGGACTATCATATGCTTACCGTAACTTGAAAGTATTTCGATTTCTTGGCTTTATATATCTTGTGGAAAGGACGAA
PCR
qPCR
Cycle Threshold (cT) are inversely proportional to the amount of target nucleic acid in the sample (ie the lower the cT = the greater the amount of target nucleic acid)
- cT = <29 = strong positive reactions / Abundant level of target primer amplicon nucleic acid (mRNA)
- cT = 30-37 = positive reactions / Moderate level of target primer amplicon nucleic acid (mRNA)
- cT = 38-40 = weak reactions / Minimum level of target primer amplicon nucleic acid (mRNA)
RFP (Red Fluorescent Protein)
RFP excites @488 nm laser/emits 585+/-8nm (similar to PE;565nm/575nm). Compatible multichannel fluorochromes with RFP = FITC, APC, PerCP-Cy5.5, PE-Cy7
- Red Fluorescent Protein Accession
- Red Fluorescent Protein Lineage
- excitation/emission (nm) 553/574
- excitation/emission (nm): 558/583
ATGAGCGAGCTGATCAAGGAGAACATGCACATGAAGCTGTACATGGAGGGCACCGTGAACAACCACCACTTCAAGTGCACATCCGAGGGCGAAGGCAAGCCCTACGAGG GCACCCAGACCATGAAGATCAAGGTGGTCGAGGGCGGCCCTCTCCCCTTCGCCTTCGACATCCTGGCTACCAGCTTCATGTACGGCAGCAAAGCCTTCATCAACCACAC CCAGGGCATCCCCGACTTCTTTAAGCAGTCCTTCCCTGAGGGCTTCACATGGGAGAGAATCACCACATACGAAGACGGGGGCGTGCTGACCGCTACCCAGGACACCAGC TTCCAGAACGGCTGCATCATCTACAACGTCAAGATCAACGGGGTGAACTTCCCATCCAACGGCCCTGTGATGCAGAAGAAAACACGCGGCTGGGAGGCCAACACCGAGA TGCTGTACCCCGCTGACGGCGGCCTGAGAGGCCACAGCCAGATGGCCCTGAAGCTCGTGGGCGGGGGCTACCTGCACTGCTCCTTCAAGACCACATACAGATCCAAGAA ACCCGCTAAGAACCTCAAGATGCCCGGCTTCCACTTCGTGGACCACAGACTGGAAAGAATCAAGGAGGCCGACAAAGAGACCTACGTCGAGCAGCACGAGATGGCTGTG GCCAAGTACTGCGACCTCCCTAGCAAACTGGGGCACAGATGA
GFP (Green Fluorescent Protein)
- Green Fluorescent Protein Accession
- Green Fluorescent Protein Lineage
- excitation/emission (nm) 482/502
ATGGAGAGCGACGAGAGCGGCCTGCCCGCCATGGAGATCGAGTGCCGCATCACCGGCACCCTGAACGGCGTGGAGTTCGAGCTGGTGGGCGGCGGAGAGGGCACCCC CAAGCAGGGCCGCATGACCAACAAGATGAAGAGCACCAAAGGCGCCCTGACCTTCAGCCCCTACCTGCTGAGCCACGTGATGGGCTACGGCTTCTACCACTTCGGCACCT ACCCCAGCGGCTACGAGAACCCCTTCCTGCACGCCATCAACAACGGCGGCTACACCAACACCCGCATCGAGAAGTACGAGGACGGCGGCGTGCTGCACGTGAGCTTCA GCTACCGCTGCGAGGCCGGCCGCGTGATCGGCGACTTCAAGGTGGTGGGCACCGGCTTCCCCGAGGACAGCGTGATCTTCACCGACAAGATCATCCGCAGCAACGCCACC GTGGAGCACCTGCACCCCATGGGCGATAACGTGCTGGTGGGCAGCTTCGCCCGCACCTTCAGCCTGCGCGACGGCGGCTACCACAGCTTCGTGGTGGACAACCACATGCAC TTCAAGAGCGCCATCCACCCCAGCATCCTGCAGAACGGGGGCCCCATGTTCGCCTTCCGCCGCGTGGAGGAGCTGCACAGCAACACCGAGCTGGGCATCGTGGAGTACCA GCACGCCTTCAAGACCCCCATCGCCTTCGCCAGATCCCGCGCTCAGTCGTCCAATTCTGCCGTGGACGGCACCGCCGGACCCGGCTCCACCGGATCTCGC
To initiate programmable DNA targeting, dCas9 coordinates RNA-duplex (crRNA+tracrRNA) scaffold. crRNA nucleotides 1-20 guide dCas9 adjacent to genomic protospacer adjacent motif(s) (PAM). Fusion of crRNA+tracrRNA generates a chimeric single guide (sgRNA).
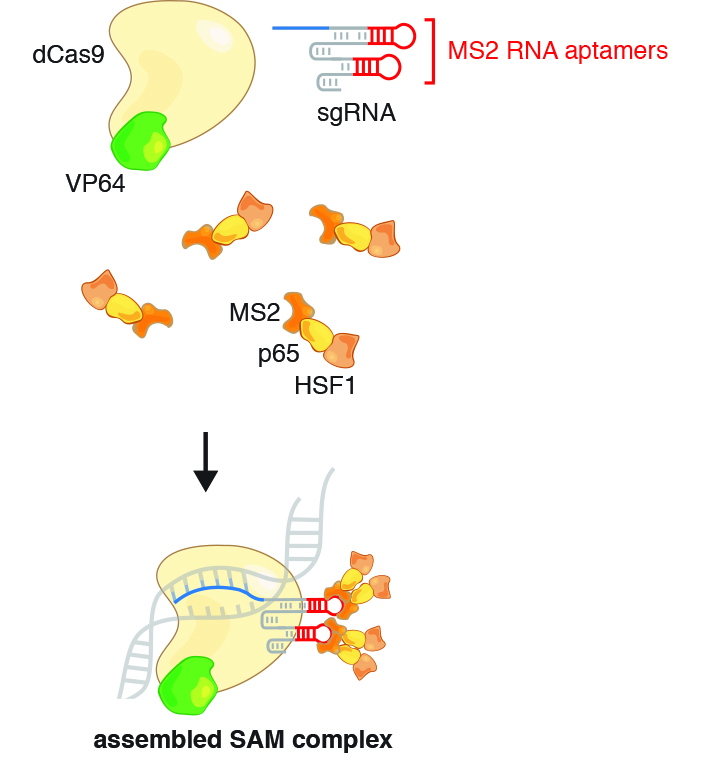
Synergistic activation mediator (SAM) system
- Incorporates two MS2 hairpins into exposed loops within the sgRNA
- Fuses MCP to a novel chimeric activator p65-HSF1 (p65 subunit of NF-kB/HSF1 heat shock factor 1 transcription response to temperature stress)
- Chimeric MCP-p65-HSF1 activators bind each MS2 hairpin as a set of homodimers
- 4 copies of MCP-p65-HSF1 scaffolding onto the SAM sgRNA, bound via dSpCas9-VP64
- Optimal upstream (-200)-(+1) >> (-400)-(-50)
Features
- Recruitment of activators in trans
- Insertion of protein binding RNA Aptamers (MS2) into sgRNA coordinates MCP
- Heterogenous activator(s) (p65-HSF1)
- Optimal upstream (-400)<->(-50) / (-200)<->(+1)
CRISPR Synergistic Activation Mediator Nomenclature
- 2A : self-cleaving peptide motif 2A peptide
- dCas9: mutant Streptococcus (pyogenes) Cas9 nuclease with inactive catalytic domains utilized as a generic RNA-guided trans-modulator. RNA-guided DNA binding protein (dCas9). dCas9 may protect internal MS2 stemloops from exonuclease degradation.
- HSF1 : Heat Shock Factor 1 transcribes gene in response to heat stress.
- MCP : MS2 Coat Protein (MCP) dimer binds each MS2 aptamer hairpin (1:1). (native MS2 system) Phage capsid assembly is nucleated by coat protein dimer binding to the operator hairpin.
- MS2 : Escherichia virus (Enterobacteria phage) MS2 RNA "operator/aptamer hairpin". (tandem repeat fusion to 3' sgRNA). noncoding scaffold. minimal hairpin aptamer selectively binds dimerized MS2 bacteriophage coat protein (MCP). MS2 stem-loop placement within the sgRNA influences transcription activation efficiency.
- p65 : NF-κB subunit. NF-κB trans-activating subunit p65 shares common co-factors with VP64, recruits a distinct subset of transcription factors and chromatin remodeling complexes.
- RTA : Human herpes virus 8 (HHV-8) transactivator.
- sgRNA: single guide RNA = chimeric Crispr RNA (crRNA) + transactivating crRNA (tracrRNA).
- VP64 : VP16 x4 / Herpes (alpha-TIF (trans-inducing factor) HHV-2 transactivator protein recruits transcription factors OCT-1 and Host cell factor 1 (HCF). (component of chimeric VP64-dCAS9).
Synergistic Activation Mediator
- The SCBT Inc. SAM complex provides a robust transcription activation system
- 1 of 3 plasmid system: CRISPR/dCas9-VP64-Blast plasmid encoding the deactivated Cas9 (dCas9)nuclease (D10A and N863A) fused to the transactivation domain VP64 + blasticidin resistance gene
- 2 of 3 plasmid system: MS2-P65-HSF1-Hygro plasmid encoding MS2-p65-HSF1 fusion protein, hygromycin resistance gene.
- 3 of 3 plasmid system: The sgRNA (MS2)-Puro plasmid encoding a target-specific 20 nt guide RNA, and a puromycin resistance gene.
Parameters
- 13ul (5,000 particle/ µL) @ 200,000 cells per 35mm diameter = MOI 0.325
- 6 Well / 10e5 (100,000 cells) per well /
200uL @5,000 particles/µL = 10e6 particles
MOI 1 = 20µL
MOI 3 = 60µL
MOI 5 = 100µL
Antibiotic Selection
- There will be a limited passage number in which a "stable" antibiotic selectable lineage can be established. The gene (mRNA) must be monitored by qPCR or other method (protein) from the culture in order to determine the durability of the knockdown/activation over passage under antibiotics.
Threshold (CRISPR activation selection)
- Puromycin : 0.5-2.0 µg/ml
- Hygromycin B : 75-150 µg/ml
- Blasticidin S HCL : 1-10 µg/ml
Upper Limit
- Puromycin : 2–10 µg/ml
- Hygromycin B : 200–500 µg/ml
- Blasticidin S HCL : 1–20 µg/ml
Empirical
- Puromycin : 2 µg/ml
- Hygromycin B : 200 µg/ml
- Blasticidin S HCL : 5 µg/ml
puromycin N-acetyltransferase
- N-acetyltransferase catalyse transfer of acetyl groups to aromatic amine (ie Tyrosine) /inactivates puromycin by acetylating amino position of tyrosine
- Puromycin Selection
- Puromycin : 0.5-2.0 µg/ml
- Puromycin : Reversible inhibitor of dipeptidyl-peptidase II (serine peptidase) and cytosol alanyl aminopeptidase (metallopeptidase). premature ribosomal peptide chain termination/ translation disruption.
ATGACCGAGTACAAGCCCACGGTGCGCCTCGCCACCCGCGACGACGTCCCCAGGGCCGTACGCACCCTCGCCGCCGCGTTCGCCGACTACCCCGCCACGCGCCACACC GTCGATCCGGACCGCCACATCGAGCGGGTCACCGAGCTGCAAGAACTCTTCCTCACGCGCGTCGGGCTCGACATCGGCAAGGTGTGGGTCGCGGACGACGGCGCCGCG GTGGCGGTCTGGACCACGCCGGAGAGCGTCGAAGCGGGGGCGGTGTTCGCCGAGATCGGCCCGCGCATGGCCGAGTTGAGCGGTTCCCGGCTGGCCGCGCAGCAACAG ATGGAAGGCCTCCTGGCGCCGCACCGGCCCAAGGAGCCCGCGTGGTTCCTGGCCACCGTCGGCGTCTCGCCCGACCACCAGGGCAAGGGTCTGGGCAGCGCCGTCGTG CTCCCCGGAGTGGAGGCGGCCGAGCGCGCCGGGGTGCCCGCCTTCCTGGAGACCTCCGCGCCCCGCAACCTCCCCTTCTACGAGCGGCTCGGCTTCACCGTCACCGCC GACGTCGAGGTGCCCGAAGGACCGCGCACCTGGTGCATGACCCGCAAGCCCGGTGCCTGA
- Streptomyces hygroscopicus produces Hygromycin B interferes with translocation/triggers mistranslation
- Phosphotransferase (HPH) modifies the aminoglycosdie antibiotic HygroB: 1026nt / ~39kD
- VERSION V01499.1
- Hygromycin B: 75-150 µg/ml (eukaryotic mammalian)
ATGAAAAAGCCTGAACTCACCGCGACGTCTGTCGAGAAGTTTCTGATCGAAAAGTTCGACAGCGTCTCCGACCTGATGCAGCTCTCGGAGGGCGAAGAATCTCGTGCTTTCAGCTTCG ATGTAGGAGGGCGTGGATATGTCCTGCGGGTAAATAGCTGCGCCGATGGTTTCTACAAAGATCGTTATGTTTATCGGCACTTTGCATCGGCCGCGCTCCCGATTCCGGAAGTGCTTGA CATTGGGGAATTCAGCGAGAGCCTGACCTATTGCATCTCCCGCCGTGCACAGGGTGTCACGTTGCAAGACCTGCCTGAAACCGAACTGCCCGCTGTTCTGCAGCCGGTCGCGGAGGCC ATGGATGCGATCGCTGCGGCCGATCTTAGCCAGACGAGCGGGTTCGGCCCATTCGGACCGCAAGGAATCGGTCAATACACTACATGGCGTGATTTCATATGCGCGATTGCTGATCCCC ATGTGTATCACTGGCAAACTGTGATGGACGACACCGTCAGTGCGTCCGTCGCGCAGGCTCTCGATGAGCTGATGCTTTGGGCCGAGGACTGCCCCGAAGTCCGGCACCTCGTGCACGC GGATTTCGGCTCCAACAATGTCCTGACGGACAATGGCCGCATAACAGCGGTCATTGACTGGAGCGAGGCGATGTTCGGGGATTCCCAATACGAGGTCGCCAACATCTTCTTCTGGAGG CCGTGGTTGGCTTGTATGGAGCAGCAGACGCGCTACTTCGAGCGGAGGCATCCGGAGCTTGCAGGATCGCCGCGGCTCCGGGCGTATATGCTCCGCATTGGTCTTGACCAACTCTATC AGAGCTTGGTTGACGGCAATTTCGATGATGCAGCTTGGGCGCAGGGTCGATGCGACGCAATCGTCCGATCCGGAGCCGGGACTGTCGGGCGTACACAAATCGCCCGCAGAAGCGCGGC CGTCTGGACCGATGGCTGTGTAGAAGTACTCGCCGATAGTGGAAACCGACGCCCCAGCACTCGTCCGAGGGCAAAGGAATAG
- Blasticidin S HCL : 1-15 µg/ml
- cytidine deaminase Zn2+ dependent Deamination of cytosine bsr
- BSR gene
- GenBank: MH238440.1 protein_id=AWG43611.1
ATGGCCAAGCCTTTGTCTCAAGAAGAATCCACCCTCATTGAAAGAGCAACGGCTACAATCAACAGCATCCCCATCTCTGAAGACTACAGCGTCGCCAGCGCAGCTCTCTCTAGCGACG GCCGCATCTTCACTGGTGTCAATGTATATCATTTTACTGGGGGACCTTGTGCAGAACTCGTGGTGCTGGGCACTGCTGCTGCTGCGGCAGCTGGCAACCTGACTTGTATCGTCGCGAT CGGAAATGAGAACAGGGGCATCTTGAGCCCCTGCGGACGGTGCCGACAGGTGCTTCTCGATCTGCATCCTGGGATCAAAGCCATAGTGAAGGACAGTGATGGACAGCCGACGGCAGTT GGGATTCGTGAATTGCTGCCCTCTGGTTATGTGTGGGAGGGCTAA
Components
【Lenti dCas9-VP64-Blast】
- dCas9 (D10A/H840A) (Streptococcus pyogenes)
- NLS: nuclear localization signal (SV40)
- VP64 (Herpes Simplex Virus derived)
- 2A peptide—>(Thosea asigna virus)
- Blastcidin resistance gene—>Bacillus cereus
- EF1a promoter—> human / versus CMV promoter, eukaryotic translation elongation factor 1 α (EF‐1α/EEF1A1) promoter increases transfection efficiency, transgene expression, and proportion of expression‐positive clones/copy number. constitutively active in a broad range of cell types
【Lenti MS2-P65-HSF1-Hygro】
- MS2:Bacteriophage MS2 coat protein
- P65:Human
- HSF1:Human
- NLS: nuclear localization signal: (SV40)
- 2A peptide:Thosea asigna virus
- Hygromycin resistance gene:E,coli
- EF1a promoter:human
【Lenti sgRNA (MS2)-Puro】
- sgRNA
- gRNA Scaffold :(MS2 aptamer-modified tetraloop and stem loop 2 (MS2 loops))
- U6 promoter:human
- Puromycin resistance gene:Streptomyces alboniger
- EF1a promoter:human
HIV derived
- 5'LTR: 5'Long Terminal Repeat (Lentivirus)
- cPPT: Central Polypurine Tract (Lentivirus)
- RRE: Rev Response Element (Lentivirus)
- Psi Sequence (Lentivirus)
- 3'LTR: 3'Long Terminal Repeat (Lentivirus)
Stem Cell / Sensitive Cell DNA Transfection
For DNA transfection of primary cells and sensitive cell lines, Effectene Transfection Reagent is a nonliposomal lipid reagent for DNA transfection into a broad range of cell types. Due to low cytotoxicity, Effectene Transfection Reagent is suitable on primary or other sensitive lineages.
Selection
- Technologies for Single-Cell Isolation
- Single Cell Isolation and Analysis
- Cloning Cylinder
- Thin layer agarose video
- Thin layer agarose method
Transgenics
Lineage Integrity
- Isogenic selection (subcloning)
Non-genetic
- (In Vivo/Ex Vivo) Transcription heterogeneity (toward antigen presentation repression, toward low clonal (proliferation) output, from/toward loss of epigenetic function) increases clonal fitness?
- Passage dependent cell-intrinsic hereditary clonal dominance.
- 3488049636108220
Intersectional genetics
- Cre recombinase: 38kDa independently acting (tyrosine) recombinase from dsDNA virus (phage) P1 bacteriophage that infects bacteria/prokaryotes. Catalyzes site specific recombination between two DNA recognition sites (LoxP (locus of X-over P1) sites; 34bp: 13 bp palindromic sequences flanking 8bp spacer).
- Floxing/Floxed/Flox (flanking/flanked by LoxP): DNA sequence flanked by two loxP sites (LoxP sandwiched DNA sequence) enables deletion (knock out) by Cre-Lox recombination.
- RMCE (recombinase-mediated cassette exchange) = modification of eukaryotic genome via targeted integration/deletion/inversion/etc.
- 38kDa Cre (integrase) recombinase: topoisomerase I-like site specific recombination between LoxP sites.
- Flp (recombinase flippase (Flp))
- type specific expression of target genes (EF1-LSL-tTA knockin) Eef1a1-2xloxP (LSL)-tetracycline-controlled transactivator (tTA). homologous recombination approach targeted insertion of (tTA) into endogenous eukaryotic translation elongation factor 1 alpha 1 (Eef1a1) locus; tTA is preceded by a transcriptional Stop sequence (i.e., three polyA signals), which is surrounded by two loxP sites (LSL).
- Dre/Cre recombinase
(Retro)Transposable elements
Recombination
- Exchange of replicating genetic information;recombination in DNA repair (ie repair of double-stranded breaks DSBs)