Gerber:What We Do
RNA-binding proteins and posttranscriptional coordination of gene expressionGene expression must be tightly controlled to ensure coordinated synthesis of the cells’ macromolecular components. Besides transcriptional control, it has become evident that also the later post-transcriptional steps – namely the processing, transport, turnover and translation of mRNAs – play pivotal roles for diversification and spatiotemporal control of gene expression. Hundreds of RNA-binding proteins (RBPs) and non-coding RNAs mediate post-transcriptional control with widespread implications in cell physiology and disease. Nevertheless, the targets and functions for most RNA-binding proteins and non-coding RNAs are not known. We are combining genome-wide analysis with classical biochemical and genetic tools to identify the RNA targets of RNA-binding proteins and to investigate post-transcriptional gene regulation at a global scale. Importantly, these studies revealed that RBPs bind to and coordinate groups of mRNAs that code for proteins, which are localized to the same subcellular compartment, act in the same pathway or are components of macromolecular complexes, forming so-called 'RNA regulons'. Moreover, these sets of RNA often bear conserved sequence/ structural elements that likely represent binding sites for RBPs. These findings suggest the presence of a highly-organized and elaborate post-transcriptional regulatory system that may affect virtually every RNA in a cell. Hence, our research mainly focuses on specific RNA-binding proteins that coordinate the localization, decay or translation of mRNAs in the cytoplasm. On the one hand, we further investigate the functions of recently discovered 'unconventional' RNA-binding proteins, such as metabolic enzymes with the aim to obtain a better understanding of the functional implications of those RNA-enzyme interactions. We further study how RBPs critically control expression of therapeutic mRNAs; and we develop models describing auto-regulatory feedback control through RNA-binding proteins. On the other hand, we characterize the 'translatome' – which refers to all mRNAs associated with ribosomes for protein synthesis (Halbeisen et al. 2009). We currently monitor translational control during ageing and sleep in the brain and under pathological conditions. Commonly, we use budding yeast as model to establish new techniques and to elucidate principles of post-transcriptional control, and we work with mammalian cells to unravel the implications in disease. 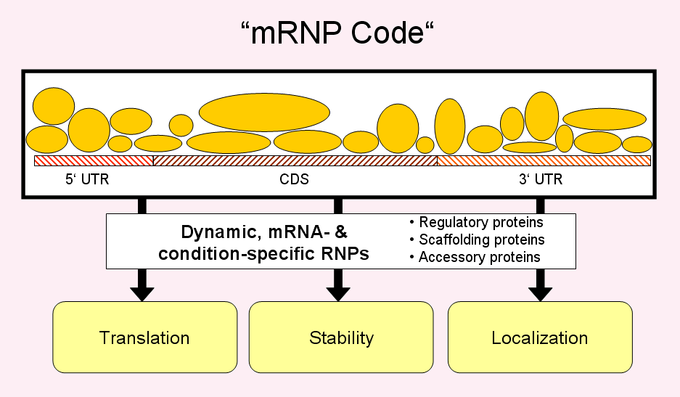 Reviews:
|