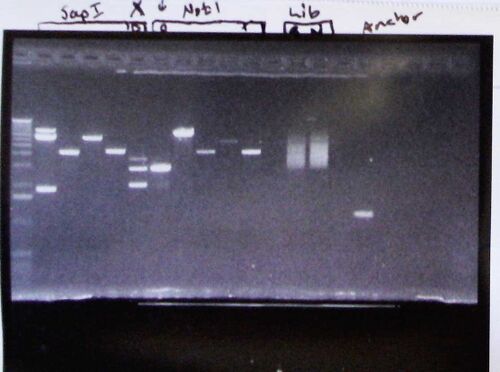
User:Anthony Salvagno/Notebook/Research/2009/05/05/Gels
From OpenWetWare
Jump to navigationJump to search
- Marker
- SapD (Steve Koch: Incomplete digestion?)
- Sap14
- Sap16
- Sap21
- SappBS (Steve Koch: Incomplete digestion?)
- NotpBS (Steve Koch: Same clone as previous lane, but complete digestion)
- NotD (Steve Koch: See lane 2; this is complete digestion with NotI as opposed to SapI)
- Not2
- Not16
- Not21
- Empty
- Saplib
- Notlib
- Empty
- Anchor
I have been naming tubes weirdly because I've been running into repetitive names. Here I have Sap# and Not# where number is the tube of which there is plasmid plus insert from a given tube (going way back) and Sap/Not is the restriction enzyme that I used to cut said tube. Sap and Notlib are libraries (after a bombing) of gDNA. See these pages:
- User:Anthony Salvagno/Notebook/Research/2009/05/01/pAS lib for library explanation (Steve Koch:I'm guessing it's double-digested EcoRI/XhoI genomic DNA, but I don't think the information was written down).
- User:Anthony Salvagno/Notebook/Research/2009/04/30/SapI Digestion of random fragments SapI digestions (lanes 2-5; not lane 6 though?)
- explanation of naming conventions
- User:Anthony Salvagno/Notebook/Research/2009/04/16/Gel Results Original shotgun clone analysis.
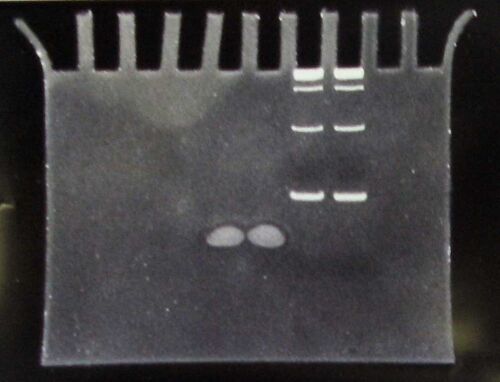
Lanes (inverted from picture):
- Marker (50bp ladder)
- Marker (50bp ladder)
- NotHP
- HP
HP is the hairpin adapter. So this gel didn't work well. I had to run this in a vertical acrylomide gel. I'll have to write the protocol up later.