Falghane Week 14/15
From OpenWetWare
Jump to navigationJump to search
Purpose
The purpose of this assignment was to model the chemostat used in Tai et al. (2007) using MATLAB.
Methods
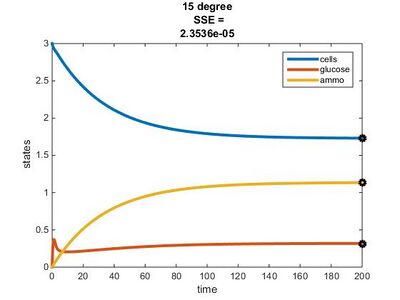
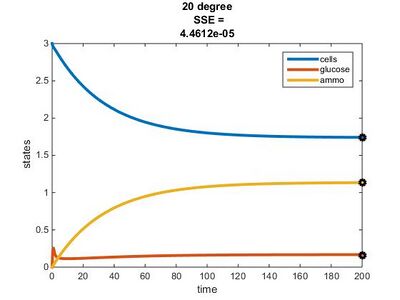
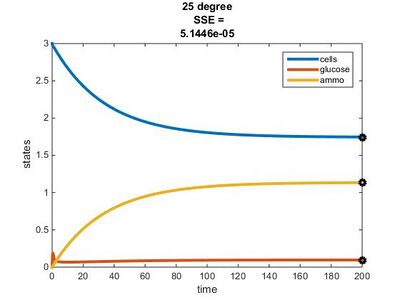
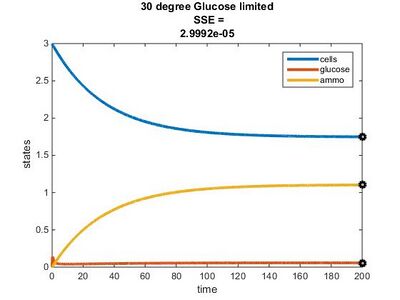
1. Use the Arrhenius equation (rate = A*exp(-B/(R*T)) to model the temperature dependence of the chemostat reaction.
- Ea was found from the slope between ln(k) and 1/temp (K) and finding B which equaled Ea.
- Equation: A = k/ e^-(Ea/RT) was then used to solve for A
- Then Arrhenius Equation: rate = A*exp(-B/(R*T ) was used to find rates at temperatures 15, 20, 25 in Kalvin.
- Then the New Rates were added to MATLAB to model temperature dependence of the chemostat reaction
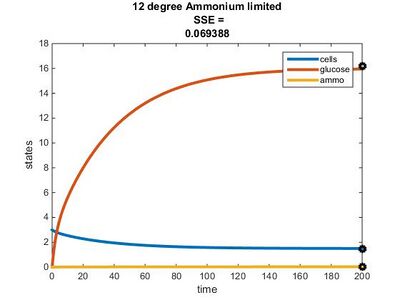
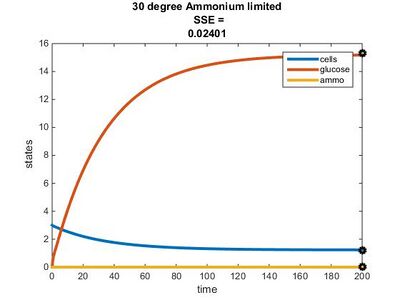
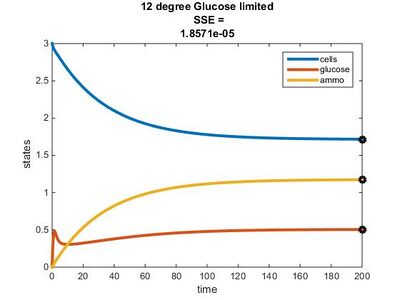
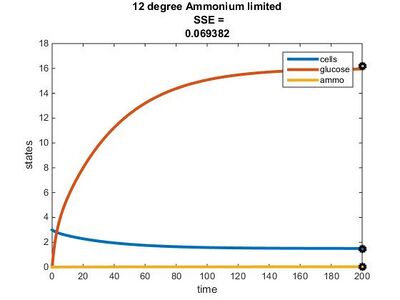
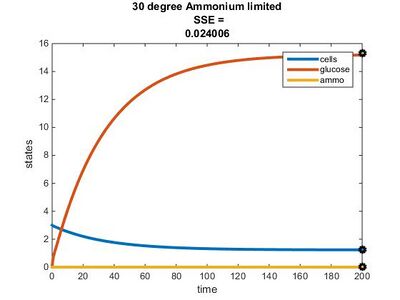
2. Investigating the glucose efficiency/ waste constant
- The values of E for glucose-limited and ammonium-limited conditions were noted.
- For each temperature (12, 30), the function E(y) that matches the two points of (y,E) data was found.
- The chemostat_2nutrient_dynamics.m file was modified to use the functions created.
- The resulting simulation was compared to the previous one.
Results
- Determing the A & B constants:
- B found to be 69,840.59 from B = (Rln(k1/k2))/(1/T1-1/T2).
- A found to be 4.979 * 10^11 from A = k/ e^-(Ea/RT).
- Rate values:
- r(15°C) = (4.979 x 10^11)e^(-69840.59)/((8.314)(288.15)) = 0.1087
- r(20°C) = (4.979 x 10^11)e^(-69840.59)/((8.314)(293.15)) = 0.1787
- r(25°C) = (4.979 x 10^11)e^(-69840.59)/((8.314)(298.15)) = 0.289
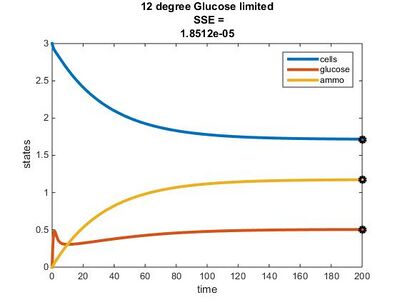
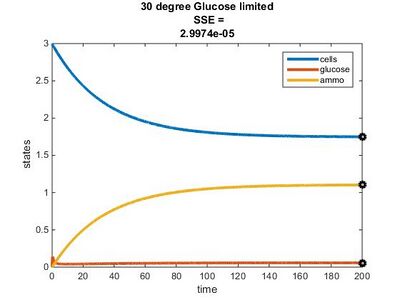
- Efficiency constant Investigation
- Original: E = 1/Y New equation: where E = my + b
- y = residual glucose
- point intercept m = (y-y)/(x-x) was used to determine m and thenew value was plugged into E = my + b to determine b
- 12°C: E = 0.363y + 14.11
- m = (14.3-20)/(0.5045-16.22) = 0.363
- b=14.11
- 30°C: E = 0.7y + 14.25
- m = (14.3-25)/(.0541-15.33) = 0.7
- b=14.25
Temperature Dependence Graphs
Efficiency Constant Graphs
Original Model
New Model
Acknowledgments
- I worked with my homework partner, Edward Ryan R. Talatala to complete this assignment.
- I also worked with Austin to work on the efficiency constant investigation.
References
- Tai, S. L., Daran-Lapujade, P., Walsh, M. C., Pronk, J. T., & Daran, J. M. (2007). Acclimation of Saccharomyces cerevisiae to low temperature: a chemostat-based transcriptome analysis. Molecular Biology of the Cell, 18(12), 5100-5112. DOI: 10.1091/mbc.e07-02-0131
- Dahlquist, K. and Fitzpatrick, B. (2019). BIOL388/S19:Week 14/15. [online] openwetware.org. Available at:Week 14/15 Assignment Page [Accessed May 8 2019].
- Tai, S. L., Boer, V. M., Daran-Lapujade, P., Walsh, M. C., de Winde, J. H., Daran, J. M., and Pronk, J. T. (2005). Two-dimensional transcriptome analysis in chemostat cultures: combinatorial effects of oxygen availability and macro- nutrient limitation in Saccharomyces cerevisiae. J. Biol. Chem. 280, 437–447.