Endy:Screening plasmid/v2.0/Design
From OpenWetWare
(Redirected from Endy:Screening plasmid 2.0/Design)
Final Design
- outline the final design and include sequence files, etc.
- be sure to outline the mutations that were included in the GFP.
Fluorescent Proteins
Design considerations
- Brighter
- This is particularly important for the RFP, which is barely detectable at low copy using SP1.0
- Codon optimization
- might enable higher expression levels
- Maturation time
- is there a maximum we require?
Experimental Setup
The choice of FPs is dependent on the experimental setup for the FACS. At MIT we have available:
- FACSAria:
- 488,633
- 407,488,633
- MOFLO
- 355,488,568/647
- 457/488,355
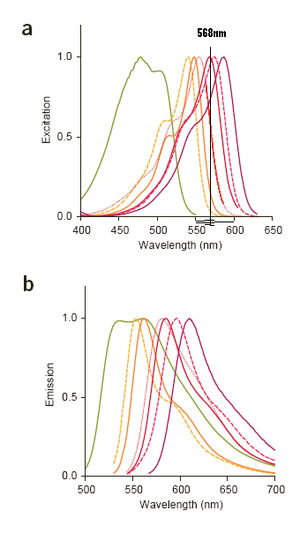
So based on the available FPs [1] it looks like the lines that matter from that list are: 407,457,488, and 568
(excitation, emission)
- 407: T-Sapphire (399,511)
- 457: Cerulean (433,475)
- 488: Emerald (487,509)
- 568: tdTomato (554,581)
Based on the brightness measurements given here[1], it looks like Emerald and tdTomato are the best choice for a compatible pair. However, there are other RFPs that could be better choices:
- (587) mCherry
- (584) J-Red
- (574) mStrawberry
- (568) LASER
- (556) dsRed
- (554) tdTomato
Frontrunners
- Emerald (GFP)
- With added A206K mutation to make it monomeric.
- tdTomato (RFP)
Codon Optimization
- Codon pairs?
- Codon bias
- Should we "optimize" for a couple different organisms if these are going to be BB's, we could do Coli and Yeast or something.
- repeat removal
- BB restrictions site removal
- Capping restrictions site removal
References
- Shaner NC, Steinbach PA, and Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005 Dec;2(12):905-9. DOI:10.1038/nmeth819 |
Inducible Expression Systems
Discussion here about inducible expression systems in E.coli.
pPRO
- Propionate-inducible induction system, has a larger dynamic range than pBAD and doesn't require a specialized strain. (i.e. no knockouts of native systems needed.)
- It looks like MC4100 is missing the propionate metabolism genes and so wouldn't work with this system, these are the two genes in K12 are homologous to the prpBCDE operon:
- 349236..350405, 350439..351890 Unfortunately, there is a 98kb deletion from the MC4100 (a K12 derivative) genome from 274723-371962, so it's missing these genes.
- However, I'm not tied to MC4100, would rather get this working in K12 and then we could maybe make some progress on doing directed deletions of K12 as planned in the Biobricks Standard Strain.
- This system is catabolite repressed by both glycerol and glucose -- What is the best minimal media + carbon source that it will work with?
- There is a version of pPro from a Salmonella that provides stronger induction. The higher expression comes along with a weaker off, so would have to make a guess if we're going to have too high expression or not. I personally think the higher the better, since eventually i would like to run this on an F plasmid or something. Though higher expression is more of a double edge sword thanks to higher "load" -- brighter FPs is really the free ride when it comes to getting better signal at low copy.
lacI w/ lacY knockout
- Pros: Use of IPTG means the inducer will not be metabolized by the cells
- Cons: Requires some strain engineering, how well do you get induction w/ IPTG w/o lacY?